Troyus fabulosus Grishin, 2023
|
publication ID |
https://doi.org/ 10.5281/zenodo.7710103 |
|
persistent identifier |
https://treatment.plazi.org/id/DD62E766-2A64-724E-FF36-C681FB3BFE6B |
|
treatment provided by |
Felipe |
|
scientific name |
Troyus fabulosus Grishin |
| status |
sp. nov. |
Troyus fabulosus Grishin , new species
https://zoobank.org/ 15B54334-E76A-44D0-853C-B15E1B423C4D
( Fig. 48 View Figure 48 part, 49, 50a, b, 51)
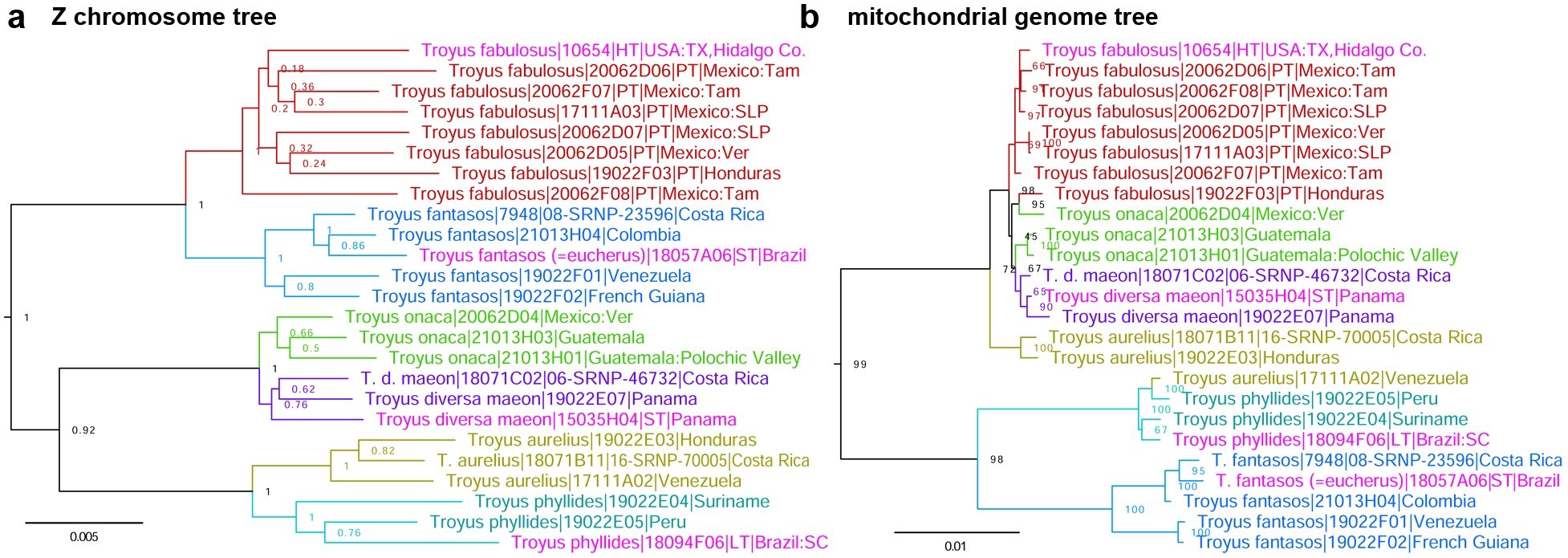
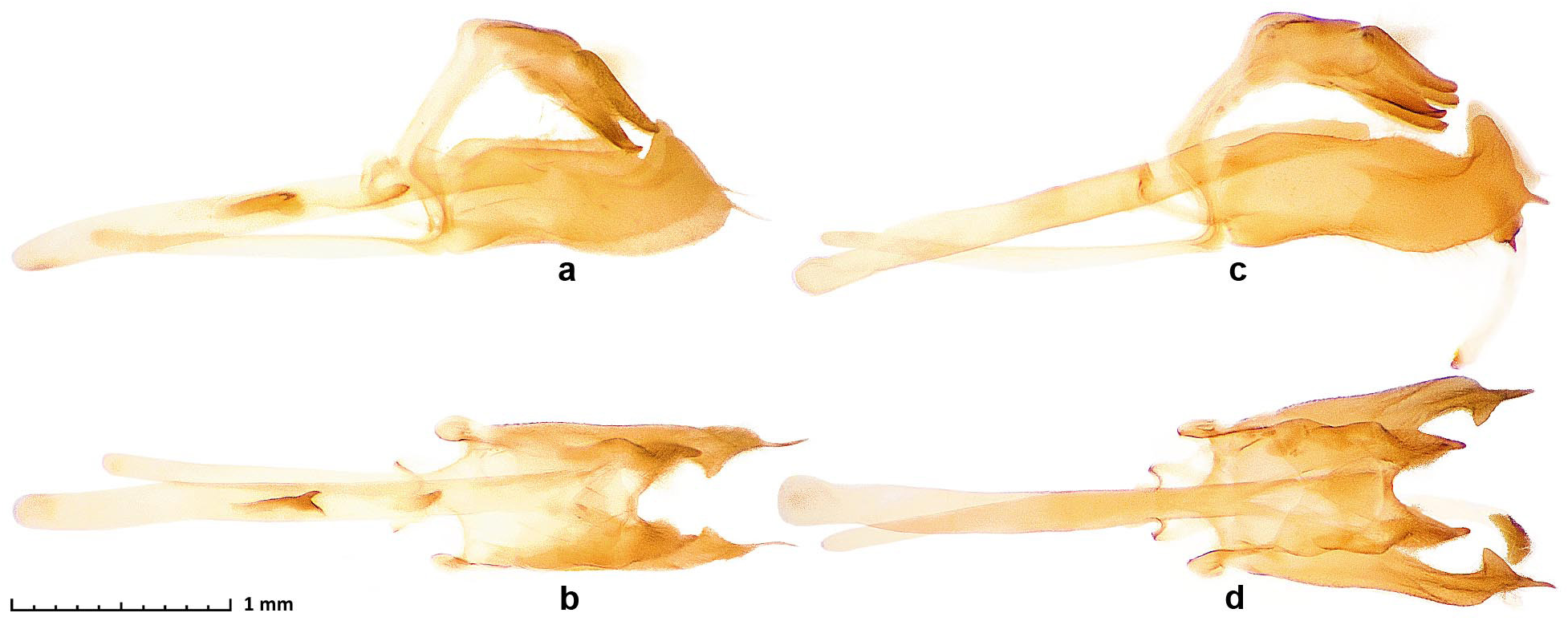
Definition and diagnosis. Inspection of genomic trees of Troyus A. Warren and Turland, 2012 (type species Troyus turneri A. Warren and Turland, 2012 ) reveals that specimens of Troyus fantasos (Cramer, 1780) (type locality in Suriname) are not monophyletic in the mitochondrial genome tree ( Fig. 48b View Figure 48 blue and red) and they are correspondingly partitioned into two clades in nuclear DNA. Fst / Gmin statistics for the comparison of the two clades are 0.38/0.006, suggesting that they represent distinct species. The blue clade contains more southern populations including a specimen from French Guiana, and these specimens agree better with the original illustration of T. fantasos , thus being this species. The red clade consists of northern populations (southernmost of those sequenced was from Honduras) and does not have a name, therefore being a new species. COI barcodes of the new species differ from those of T. fantasos by 4.3% (28 bp). However, it shares the barcodes (and the rest of mitochondrial DNA, Fig. 48b View Figure 48 ) with sympatric Troyus onaca ( Evans, 1955) and Troyus diversa maeon (Mabille, 1891) . Moreover, we see mitochondrial DNA introgression between T. aurelius (Plötz, 1882) ( Fig. 48b View Figure 48 olive) and T. phyllides (Röber, 1925) ( Fig. 48b View Figure 48 cyan), suggesting evolutionary peculiarities in this genus and underscoring the importance of nuclear DNA analysis ( Fig. 48a View Figure 48 ). The new species keys to “ Vettius fantasos fantasos ” (J.45.12(b)) in Evans (1955), and differs from it by generally paler and less contrasting ventral hindwing: area between veins M 1 and M 3 that is mostly pale distad of the postdiscal rust-colored cross-bar ( Fig. 49 View Figure 49 , 50a, b View Figure 50 ), but is dark in typical T. fantasos ( Fig. 50c, d View Figure 50 ); the cross-bar is usually more distal in T. fantasos than in the new species, making the postdiscal pale spot appear smaller in the new species than in T. fantasos ; the dorsal hindwing pale spots are whiter than more typically brownish-orange spots of T. fantasos ; the valva is broader (in lateral view), the harpe is less robust and its posterior spike-like projection is thinner ( Fig. 51a,b View Figure 51 ) compared to T. fantasos ( Fig. 51c,d View Figure 51 ). In DNA, a combination of the following base pairs is diagnostic in nuclear genome: aly528.39.5:A75G, aly 1409.3.1:A174T, aly669.14.1:A672T, aly536.145.4:G198A, and aly 2085.2.4:T943C, and COI barcode differs from the phenotypically similar T. fantasos : C49C(not T), C82C(not T), C121C(not T), C145C(not T), and C343C(not A), but is shared with the phenotypically distinct T. onaca and T. diversa maeon .
Barcode sequence of the holotype. Sample NVG-10654, GenBank OP762113, 658 base pairs: AACTTTATACTTTATTTTTGGAATTTGAGCAGGAATACTAGGAACATCCTTAAGTTTATTAATTCGAACAGAATTAGGTAACCCTGGAT CTTTAATTGGAGATGATCAAATTTATAATACCATTGTAACAGCTCATGCTTTTATCATAATTTTTTTTATAGTTATACCAATTATAATTG GAGGATTTGGAAATTGATTAGTCCCATTAATATTAGGAGCTCCCGATATAGCATTCCCACGAATAAATAATATAAGATTTTGAATATTAC CTCCTTCTTTAATACTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACAGTTTACCCCCCTCTTTCATCTAA TATTGCCCACCAAGGAGCTTCTGTTGATTTAGCAATTTTTTCTCTTCATTTAGCTGGGATTTCATCAATTCTAGGAGCAATTAATTTTA TTACTACAATCATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGTATTACAGCATTATTATT ACTTTTATCACTACCGGTATTAGCAGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAACACATCTTTTTTTGATCCTGCTGGAGG AGGAGATCCAATCTTATATCAACATTTATTT
Type material. Holotype: ♂ deposited in the Texas A&M University Insect Collection, College Station, Texas, USA ( TAMU), illustrated in Fig. 49 View Figure 49 , bears the following seven rectangular labels, six white: [TEXAS: | HIDALGO COUNTY | Penitas], [coll. | 24-Oct-1975 | Edward C. Knudson], [Allyn Museum photo | No. 810610 7-8], [ HESPERIIDAE , | Hesperiinae : | Vettius fantasos | (Stoll, [1780]) | ♂ det. R.O. Kendall | [M. & B. No. 137.5]], [DNA sample ID: | NVG-10654 | c/o Nick V. Grishin], [genitalia | NVG180106-71 | Nick V. Grishin], and one red [HOLOTYPE ♂ | Troyus fabulosus | Grishin]. Paratypes: 7♂♂ in TMMC unless specified otherwise: Mexico: Tamaulipas: 1♂ NVG-20062D06 1–4 km N of Gomez Farias, 350 m, 10-Aug-1981, C. J. Durden; 2♂ NVG-20062F07 and NVG-20062F08, 13 km NNE of Chamal, 1600 ft, 13-Jan-1969; San Luis Potosi: 1♂ NVG- 20062D07 Maiz, El Sabinito, 14-Jun-1979, C. J. Durden; 1♂ NVG-17111A03 El Platanito, 24-Jun-1983, W. H. Howe leg. [ LACM]; 1♂ NVG-20062D05 Veracruz, 4.5 km SW of Omealca, 7-Aug-1981, C. J. Durden; Honduras 1♂ NVG-19022F03 San Pedro Sula, 27-Dec-1978, Robert D. Lehman leg. [ USNM].
Type locality. USA: Texas, Hidalgo Co., Peñitas.
Etymology. The name implies that this northern counterpart of South American T. fantasos is no less fantastic than it. The name is a masculine adjective.
English name. Fabulous skipper.
Distribution. Currently known from South Texas, USA, Mexico, and Honduras.
Comment. Incongruence between nuclear and mitochondrial DNA trees ( Fig. 48a View Figure 48 vs. b) is remarkable in Troyus . It highlights the dangers of relying exclusively on mitochondrial DNA in phylogenetic studies.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |