Limnephilus spp.
|
publication ID |
https://doi.org/ 10.1093/isd/ixab013 |
|
persistent identifier |
https://treatment.plazi.org/id/03C887EB-FFDE-521B-FF23-11A816988208 |
|
treatment provided by |
Felipe |
|
scientific name |
Limnephilus spp. |
| status |
|
The morphological characters were well in line with the literature and we found no clear indications of cryptic species. Only minor differences in the shapes and setae arrangement of genital segments and their appendages were detected, whereas species-level differences between North European Limnephilids are regularly clearly more significant ( Malicky 2004). Also, when examining L. sericeus adults, we found no characters of closely related North American L. sericeus group of species ( Ruiter 1995).
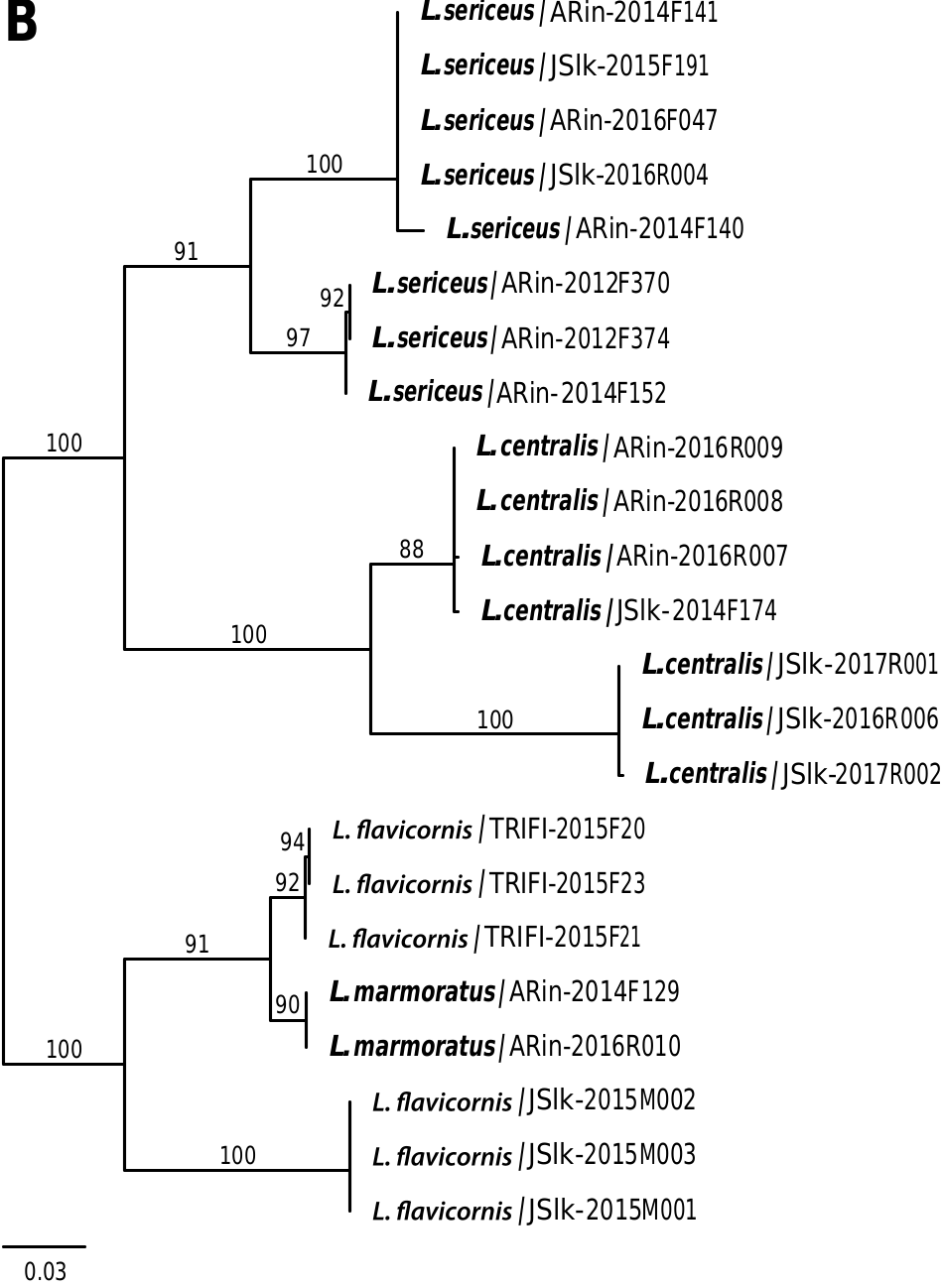
The samples of each three Limnephilus species were split into two DNA barcode clusters: (i) Limnephilus sericeus with mean Kimura-2 parameter (K2P) divergence of 6.8% between the clusters, and mean intra-cluster K2P divergence of 0.3%; (ii) Limnephilus centralis with mean K2P divergence of 8.8% between the clusters and mean intra-cluster K2P divergence of 0.1%; and (iii) Limnephilus flavicornis with mean K2P divergence of 10.1% between the clusters, and mean intra-cluster K2P divergence of 0.1%. One of L. flavicornis clusters is closer (mean K2P divergence 2.2%) to the DNA barcodes of a related species L. marmoratus than the second cluster of L. flavicornis .
K =7 K =6 K =4
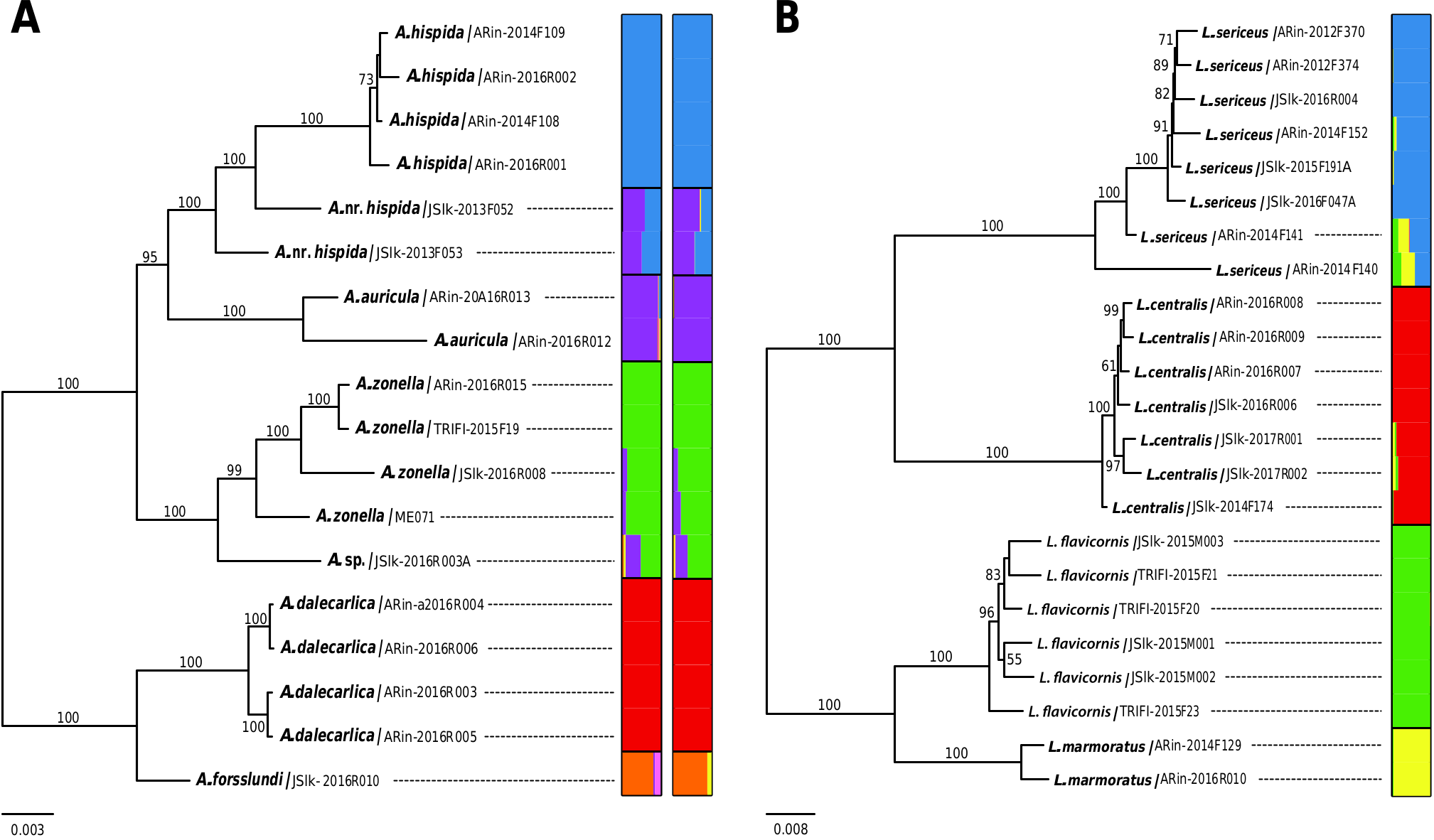
For ddRAD data of Limnephilus from 23 individuals, an average of 2.3 million raw reads was obtained (Supp Table 1 [online only]). We recovered a total of ca. 3.4 million base pairs with 20,003 loci and 81,701 SNPs, with average cluster depth of 207 ( Table 3; Supp Table 1 [online only]). The proportion of missing data was 72.7% across all loci.
Pairwise distances of ddRAD data were consistent with the current species taxonomy. Mean within species divergence varied from 0.38% ( L. centralis ) to 0.95% ( L. sericeus ), whereas distances among species were more than three times higher, i.e., from 2.86 (between L. flavicornis and L. marmoratus ) to 5.26% (between L. flavicornis and L. sericeus ).
In the mtCOI ML tree, three Limnephilus species ( L. centralis , L. sericeus , and L. flavicornis ) showed a deep intraspecific split. Two samples of L. marmoratus were sister and closely related to one of the L. flavicornis clades ( Fig. 2B View Fig ). In the ddRAD ML tree, all four species of Limnephilus were fully supported as monophyletic clades ( Fig. 3B View Fig ). The phylogenetic tree analyses supported L. flavicornis as a sister species to L. marmoratus .
The STRUCTURE identified four clusters ( Fig. 3B View Fig ), which are perfectly compatible with the ddRAD ML tree. Some signs of admixture were observed between a few heterospecific samples. Interestingly, two samples (ARin-2014F140 and ARin-2014F141) of L. sericeus showed some admixture with both L. flavicornis and L. marmoratus . Tests of admixture using D-statistics confirmed the significant levels of gene flow between these two species (Supp Table 3 [online only]).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |