Oarisma ( Oarisma ) castanea (O. Mielke, 1969), 2023
|
publication ID |
https://doi.org/10.11646/zootaxa.5271.1.3 |
|
publication LSID |
lsid:zoobank.org:pub:39D641B7-1800-4918-8E88-4EC5FF4BB56C |
|
DOI |
https://doi.org/10.5281/zenodo.7864348 |
|
persistent identifier |
https://treatment.plazi.org/id/F84A87F4-9B25-FFD0-FF3C-A172BE1DF875 |
|
treatment provided by |
Plazi |
|
scientific name |
Oarisma ( Oarisma ) castanea (O. Mielke, 1969) |
| status |
comb. nov. |
Oarisma ( Oarisma) castanea (O. Mielke, 1969) , comb. nov. originates in deep radiation of Oarisma Scudder, 1872
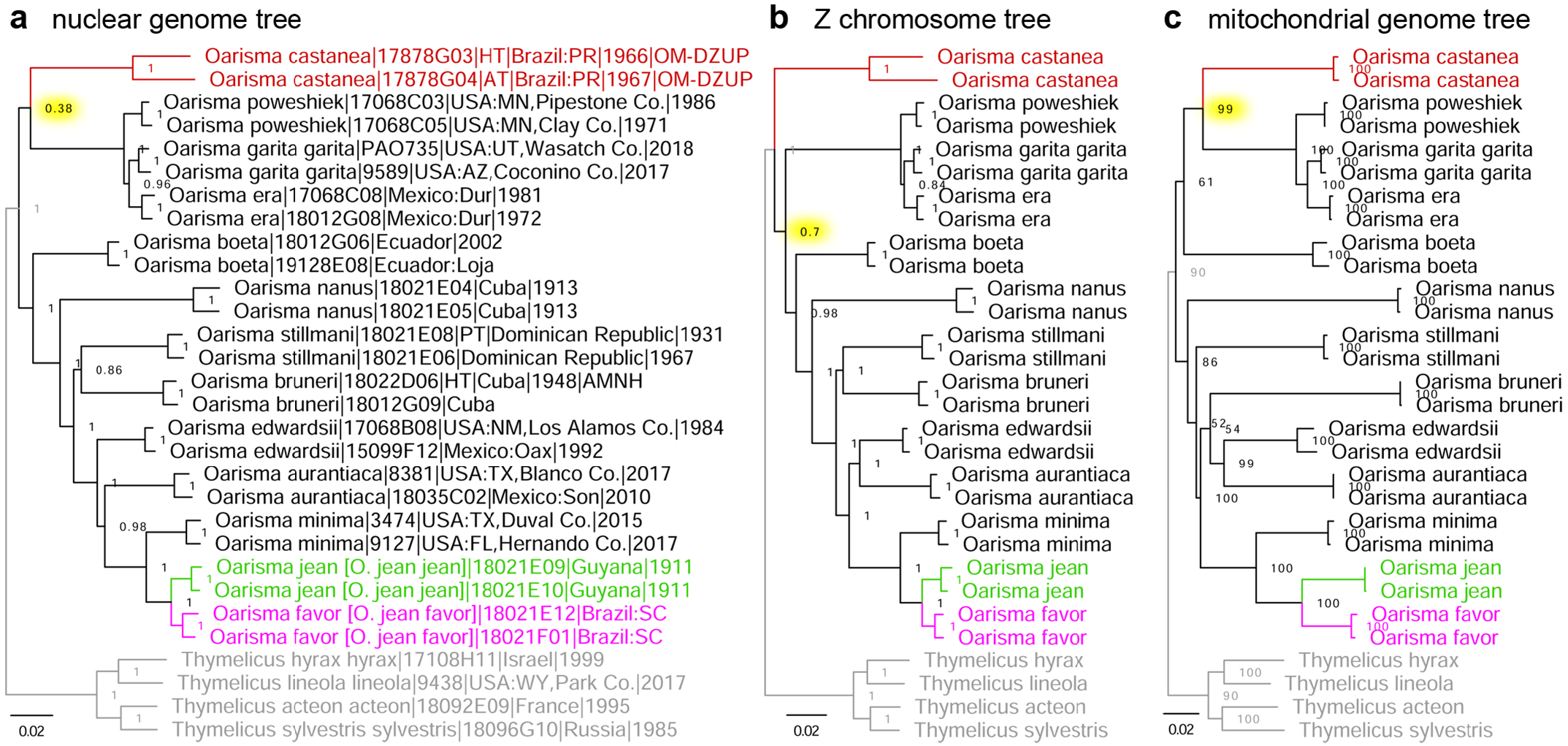
Phylogenetic analysis of the holotype and allotype of Copaeodes castanea O. Mielke, 1969 (type locality in Brazil: Paraná), presently placed in the subgenus Copaeodes Speyer, 1877 of Oarisma Scudder, 1872 (type species Hesperia powesheik [sic!] Parker, 1870), reveals that while its monophyly with Oarisma is strongly supported in all three trees (nuclear, Z chromosome and mitochondrial), its position with the genus is not well defined ( Fig. 9 View FIGURE 9 red). In the tree constructed from protein-coding regions in the Z chromosome (diverges readily in speciation and more resistant to gene exchange between species), O. castanea . is sister to other Oarisma with moderate support: 70% of position partitions yield this topology ( Fig. 9b View FIGURE 9 ). However, in the trees constructed from protein coding regions in the rest of the nuclear genome (excluding the Z chromosome) ( Fig. 9a View FIGURE 9 ) and in mitochondrial genome ( Fig. 9c View FIGURE 9 ), it is sister to Oarisma sensu stricto, weakly supported (38% of partitions) in nuclear genome and strongly supported (99% of partitions) in mitogenome. Because the number of positions included in the analysis is very large, poor statistical support is not a consequence of insufficient data. As usual with genomic data, poor support indicates rapid radiation at some point in the past and suggests incomplete lineage sorting and/or gene exchange near the origins of these species. While evolutionary processes are being investigated, we propose to classify this species according to the tree that received strongest support, i.e., mitogenome-based (99%, highlighted yellow in Fig 9c View FIGURE 9 ), and tentatively place O. castanea in the subgenus Oarisma . Furthermore, the trees reveal that the last common ancestor of Oarisma rapidly diverged into 4 lineages: subgenus Oarisma , O. castanea , O. boeta (Hewitson, 1870) , and the rest of the genus. Topologies of the three trees are inconsistent in placing the root among these four lineages ( Fig. 9 View FIGURE 9 ). This rapid radiation of Oarisma into four lineages with unresolved topology supports the treatment of these species as congeneric, because partitions into clades within this genus are not well defined.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
SubTribe |
Thymelicina |
|
Genus |