Ganoderma sinense, J. D. Zhao, L. W. Hsu & X. Q. Zhang, J. D. Zhao, L. W. Hsu & X. Q. Zhang
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2020.112466 |
|
DOI |
https://doi.org/10.5281/zenodo.8305138 |
|
persistent identifier |
https://treatment.plazi.org/id/BB74B629-FFE1-FFCC-E651-FB9904586981 |
|
treatment provided by |
Felipe |
|
scientific name |
Ganoderma sinense |
| status |
|
2.1. Prediction of anticancer activity of G. sinense View in CoL View at ENA proteins
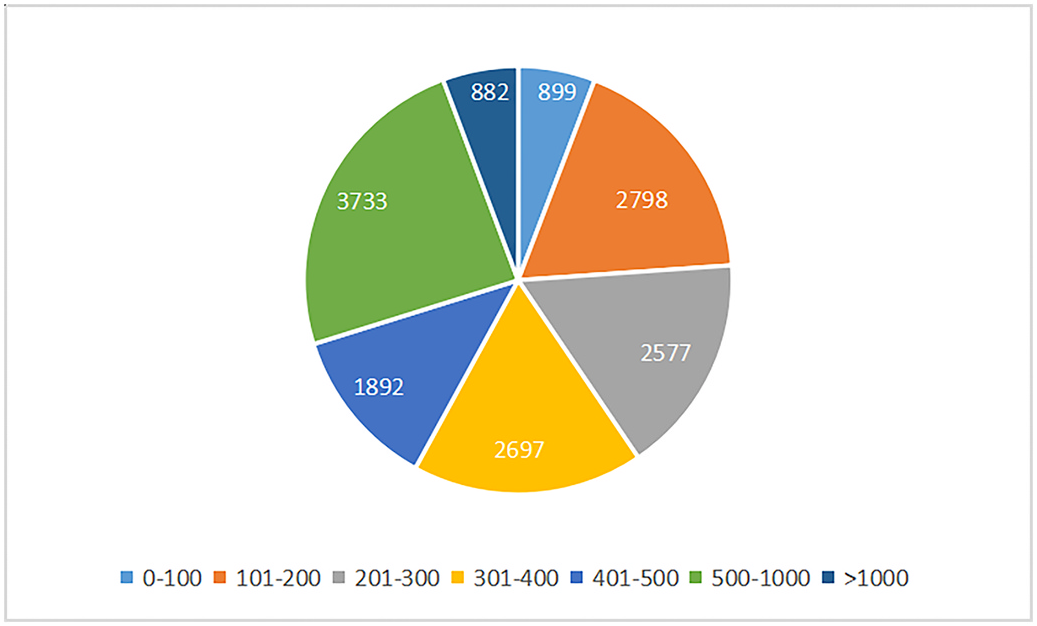
Genomic and proteomic data on G. sinense were obtained from NCBI genomic database, and include over 15,000 predicted protein sequences ( Zhu et al., 2015). The length distribution of these proteins is shown in Fig. 1 View Fig . Most of the translated proteins have more than 200 amino acid residues, and only 5.8% proteins (899 sequences) are shorter than 100 amino acids in length. First, we attempted to identify ACPs from the proteome of G. sinense ; thus, proteins shorter than 100 amino acids in length were screened by mACPpred. None of the proteins translated from the G. sinense genome was predicted to have anticancer activity. It is possible that bioactive peptides are in fact present but are inactive in their parent proteins, exhibiting activity only after being cleaved by enzymes (Manzanares et al., 2015). Considering that Ganoderma spp. is consumed as a health-care food and medicine, trypsin was selected as a model enzyme to hydrolyze the proteins. The proteins were blasted with a dataset containing 760 known ACPs to investigate whether they contain ACP sequences ( Chen et al., 2016; Wei et al., 2018; Tyagi et al., 2013). The proteins containing more than four ACP sequences were then in silico digested by trypsin, and the resulting fragments were screened by mACPpred to identify possible ACPs. A flowchart of the overall procedure is shown in Fig. 2. View Fig
2.2. Prediction of ACP-containing parent proteins
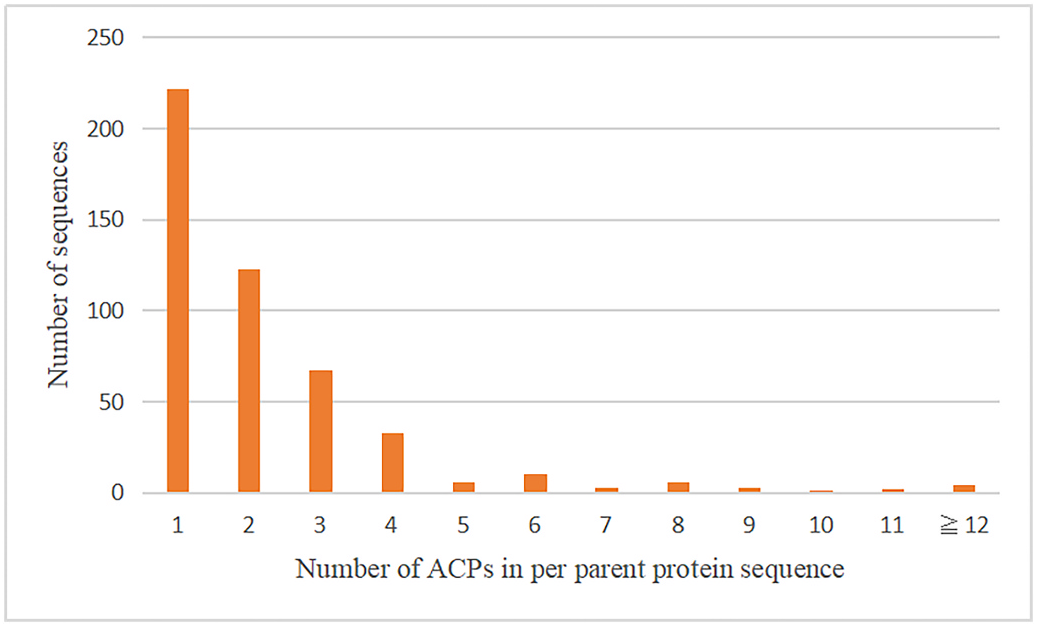
The sequences of G. sinense proteins comprising fewer than 100 amino acids were blasted against a dataset of known ACPs. The sequence alignment results are shown in Fig. 3. A View Fig total of 477 sequences possessed at least one possible ACP sequence. Some of these sequences from the G. sinense proteome (parent proteins) were similar to one or more anticancer peptide sequences. Some of the parent proteins possess multiple anticancer peptides; 26 parent proteins contained more than 6 putative ACP sequences, 4 of which contain more than 12 possible ACPs ( Fig. 3 View Fig ).
2.3. In silico enzyme digestion and ACP screening of digested fragments
Most of the currently discovered active peptides are hydrolysates of proteins. Many of the products play regulatory roles and exhibit a range of activities, including antimicrobial, anticancer, antihypertensive and immunomodulatory activities ( Yousefi et al., 2017; Manzanares et al., 2015). Trypsin was used as a model enzyme to hydrolyze parent proteins from G. sinense . The parent proteins containing more than four putative ACP sequences (a total of 65 proteins) were subjected to in silico trypsin digestion. The fragments generated by trypsin cleavage are listed in the supplement ( Table S1 View Table 1 ).
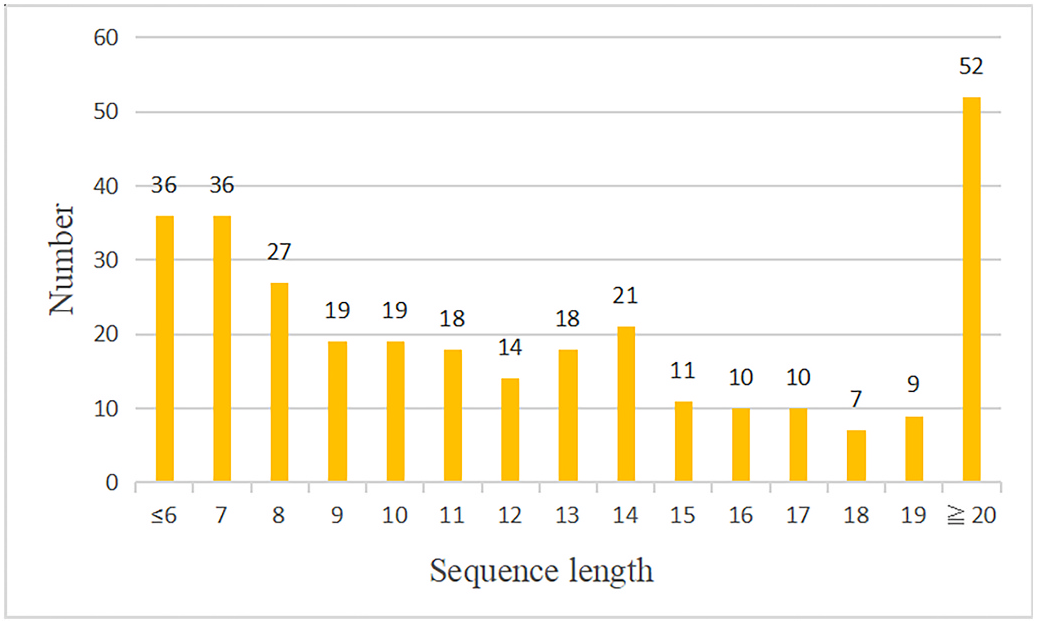
After trypsin cleavage, a total of 307 peptides were produced. The length distribution is shown in Fig. 4 View Fig mACPpred was used to predict the anticancer activity of these cleaved peptides. The results are shown in Table 1. A View Table 1 total of 34 trypsin-cleaved G. sinense proteins were predicted to produce peptides with anticancer activity. In addition, some parent proteins were predicted to generate multiple fragments with anticancer activity. These ACPs might serve as active components of orally administered G. sinense as an anticancer drug.
2.4. Verification of the screened ACPs
To verify the anticancer activity of the mACPpred-screened ACPs, the peptides were blasted against known peptides with anticancer activity. Fifteen cleavage peptides had similar sequences to those of known anticancer peptides. These cleavage peptides belong to 14 parent proteins of G. sinense ( Table 2 View Table 2 ).
According to the results, the trypsin cleavage fragments of protein No. 11 (with proteins numbered according to their order in the proteome) and No. 20 produced the same sequences as known anticancer peptides (Identity =100%). Protein No. 2 yielded two peptide hits, with one of the peptides being identical in sequence to a known anticancer peptide. The results suggested that after oral administration of G. sinense , the parent proteins of G. sinense may be digested by trypsin, resulting in the release of ACPs. Jeong et al. reported that the anticancer peptide lunasin, a 43-amino acid peptide naturally present in soybean, was bioactive and presented in liver, kidney and blood after feeding rye to rats ( Jeong et al., 2009). The bioavailability of ACPs resulting from the digestion of G. sinense proteins needs further study. Our results indicated that future research should perform in vitro digestion to aid the discovery of bioactive peptides from G. sinense or other TCMs.
2.5. Structural simulation of anticancer peptides based on molecular dynamics
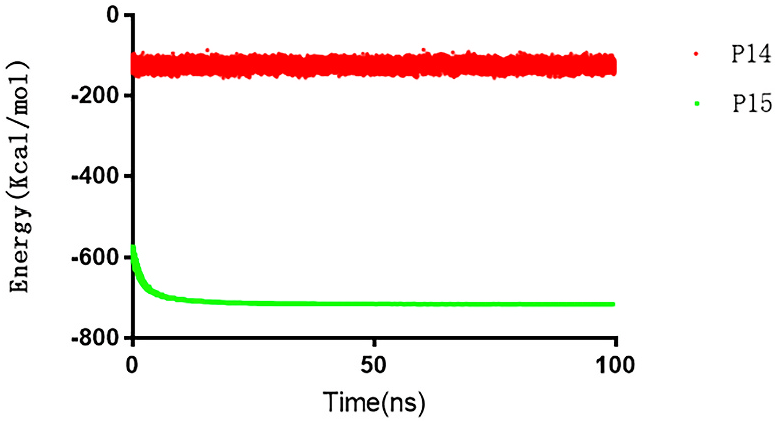
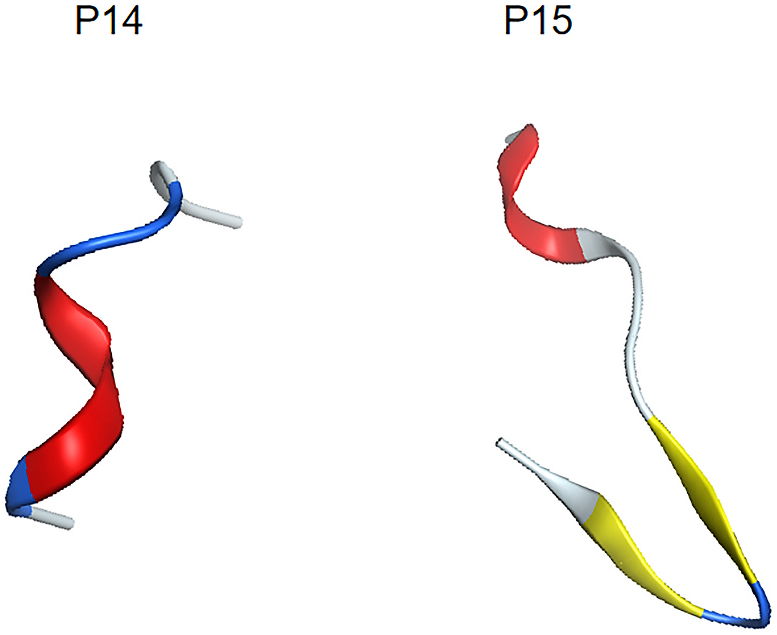
G. sinense protein Nos. 14 and 15 produced fragments similar to multiple known anticancer peptides. This result indicated that these peptides have a high probability of being unreported ACPs. Molecular dynamics were used to construct structure models of these peptides to gain insight into their possible anticancer mechanisms; these models may aid the design of novel anticancer peptides. The energy changes of these peptides after 100 ns of molecular dynamics simulation are shown in Fig. 5 View Fig . During the equilibration phase, a brief energy change is experienced, followed by an energy minimize approach to stabilization. Therefore, it can be inferred that these peptides were in a relatively stable state after a period of stretching. We extracted the lowest energy conformation from 20 to 100 ns for analysis. The results are shown in Fig. 6. View Fig
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.