Erythrophleum
|
publication ID |
https://doi.org/ 10.1071/SB23007 |
|
persistent identifier |
https://treatment.plazi.org/id/BB4F8763-FFD6-FFA8-FCB2-F9FCFD07FDC0 |
|
treatment provided by |
Felipe |
|
scientific name |
Erythrophleum |
| status |
|
Phylogeny of Erythrophleum View in CoL
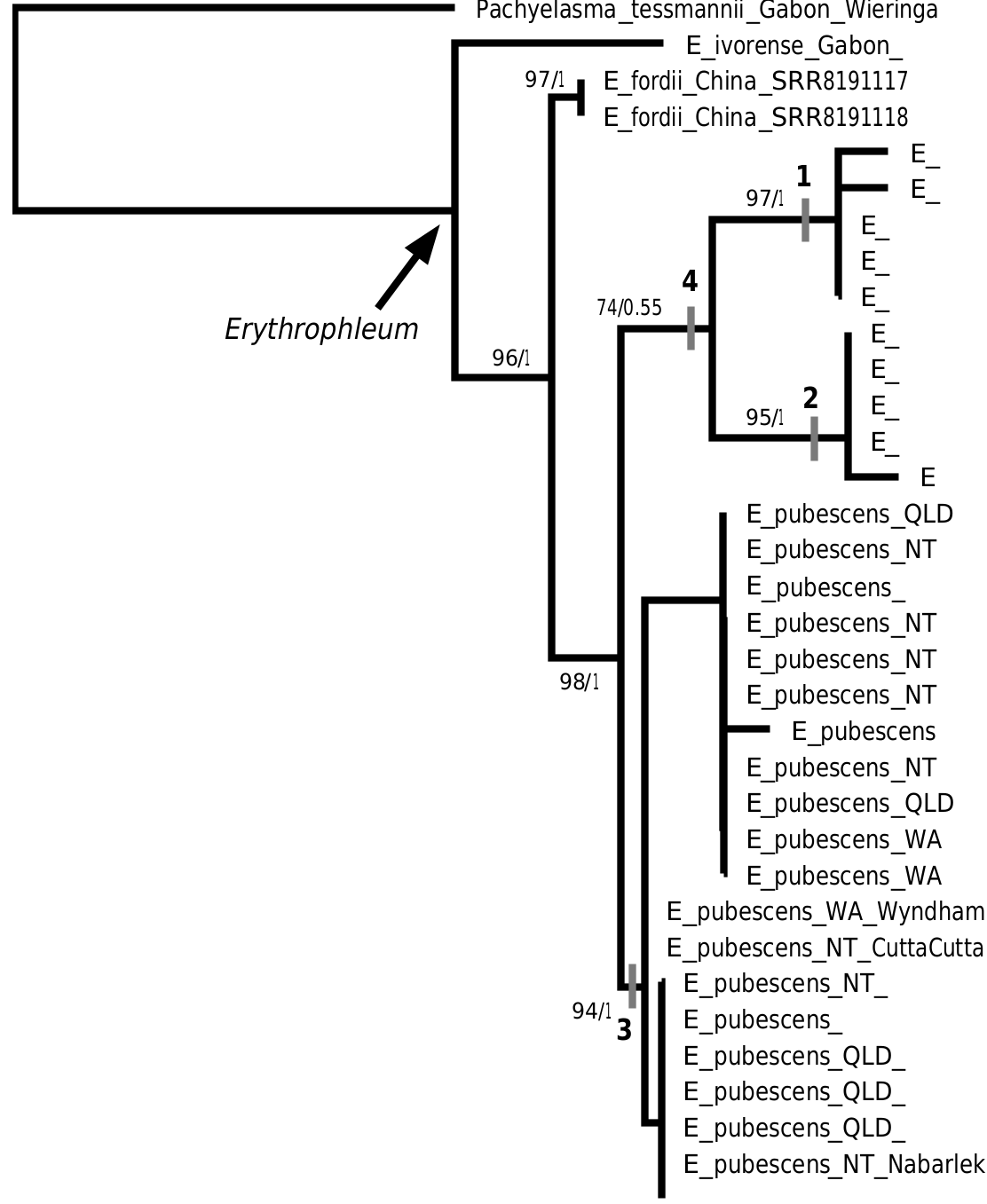
Although only two non-Australian Erythrophleum species were represented in the ribosomal tree, the Australian clade was strongly supported as sister to the Asian species E. fordii , as recovered in chloroplast phylogenies ( Duminil et al. 2013, 2015). Denser sampling of African Erythrophleum is required to further elucidate relationships and biogeography of the genus.
Although most samples used for this study were represented by ITS1 only, careful phylogenetic selection of full-length ribosomal sequences enabled robust phylogenetic
5229_ERR4363236 Wieringa5487_ERR4363217
arenarium_ WA _DampierDowns_Barrett9080_ MT 581272 arenarium_ WA _ShayGap_Bean25079_ MT 581273 arenarium_ WA _Pardoo_Sweedman8997_ MT 581271 E. arenarium arenarium_ WA _AnnaPlains_Byrne1271_ MT 581270 arenarium_ WA _DampierDowns_Forbes2465_ MT 581269 chlorostachys_ NT _FishRiver_Larcombe2_ MT 581277 chlorostachys_ NT _NitmilukNP_Wightman5205_ MT 581278 chlorostachys_ WA _LakeArgyle_Weston12284_ MT 581274 E. chlorostachys chlorostachys_ WA _KingRiver_Byrne3693_ MT 581275 _chlorostachys _ NT _ Kakadu _Lazarides8845_ MT 581276 _KingsPlainsStn_ McDonald 17554_Copy2_OQ396764 _Darwin_Smith128_ MT 581296 DrysdaleRiverMouth_Barrett5902_1_ MT 581285 _LitchfieldNP_Cowie5303_ MT 581292 _StuartHwy_Whaite3979_ MT 581283 _DungowanStn_Brennan4583_ MT 581290 _ WA _CharnleyRiverStn_DaunceyH666_ MT 581297 _BlackJungle_Smith101_ MT 581295 _Cooktown_Blake23184_ MT 581281 _KuriBay_Foulkes340_ MT 581280 E. pubescens _DunhemR_Coate224_ MT 581279 _Byrne3721_ MT 581282 _Egan2873_ MT 581294 WalkerRiver_Clark1670_ MT 581291 DrysdaleRiverMouth_Barrett5902_2_ MT 581286 Mareeba_Wannan213_ MT 581284 CapeYork_ McDonald 9767_ MT 581287 Townsville_LeitchQDA003815_ MT 581288 _Dunlop7145_ MT 581293 E_chlorostachys _QLD_KingsPlainsStn_ McDonald 17554_Copy1_OQ471964
0.007
reconstruction. Because all Australian samples were recovered as a well-supported monophyletic Australian clade ( Fig. 1 View Fig ), the sampled taxon–locus combination enabled the full-length ribosomal sequences to resolve relationships of the Australian clade to the outgroups, for which ITS1 alone proved too variable to resolve with high support (data not shown), whereas the ITS1 region alone was sufficient to resolve relationships within the more densely sampled Australian clade, with high support values.
The division of Australian Erythrophleum into three species was supported by the moderately to well-supported clustering of ITS1 sequences into three discrete clades, matching morphological designations. The greatest intraspecific variation in ITS1 sequences was found within the most widely distributed species, E. pubescens ; however, clades within this species were not supported by maximum-likelihood bootstrap or Bayesian analyses, and there is no geographical trend to the clades. Indeed, two plants from the same population (Drysdale River Mouth, Barrett MDB 5902) were placed divergently, one each in the two most numerous clades. Similarly, two ITS assemblages representing each clade were reconstructed from the single available E. pubescens short-read archive, indicating that these copies can both be present within individuals; it is possible that this is a fixed polymorphism in E. pubescens , through duplication of the ribosomal locus. Consequently, we interpret both variants as intraspecific variation within a single species, E. pubescens .
Within the three Australian species, E. arenarium and E. chlorostachys were recovered as sister taxa in the RAxML tree (74% BS), but not in the MrBayes tree (0.55 PP). This node was supported by a diagnostic 3 - bp ( GAA) insertion in the ITS1 alignment (Indel 4, Fig. 1 View Fig ), as described under results, providing additional evidence for this sister relationship in the ribosomal phylogeny. The sister relationship of these two species is a plausible result, with the two species sharing (relative to E. pubescens ) non-corky branchlets, fewer leaflets per pinna, not or slightly asymmetric leaflets, glabrous raceme axis and pedicels, and longer calyx, anthers and pod stipe, and generally shorter aril, which are possible apomorphies for the clade. However further multi-locus data are required to conclusively demonstrate this relationship.
| WA |
University of Warsaw |
| MT |
Mus. Tinro, Vladyvostok |
| NT |
Department of Natural Resources, Environment and the Arts |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |