Barbarea vulgaris, W. T. Aiton.
|
publication ID |
https://doi.org/10.1016/j.phytochem.2021.112658 |
|
DOI |
https://doi.org/10.5281/zenodo.8273670 |
|
persistent identifier |
https://treatment.plazi.org/id/66798798-FFA3-FFFB-634C-F96BFA8EF8F0 |
|
treatment provided by |
Felipe |
|
scientific name |
Barbarea vulgaris |
| status |
|
2.2.7. A search for aliphatic glucosinolates in Barbarea vulgaris View in CoL View at ENA
As observed for the Australian Barbarea species (Section 2.2.1.), aliphatic GSLs generally appear to be absent from the genus, except for a single historical report of isothiocyanate (ITC) hydrolysis products of aliphatic GSLs, prop-2-enyl ITC, isopropyl ITC and 3-(methylthio)propyl ITC, from Barbarea spp. ( Cole, 1976) . However, identification of some ITCs in that report was of questionable reliability ( Olsen et al., 2016). Indeed, that historical paper also (unexpectedly) reported prop-2-enyl
ITC from Plantago major L., ( Plantaginaceae : Lamiales ] although competent researchers were later unable to detect any GSL in that species ( Larsen et al., 1983). However, more recently data also suggested the possibility of aliphatic GSLs in Barbarea . Three observations were particularly suggestive: CYP79F6 and other putative GSL biosynthetic enzymes were reported to be induced by larvae of the moth Plutella xylostella (L.) ( Liu et al., 2016), B. vulgaris amino acid chain elongating enzymes were reported to chain elongate Met and Leu in addition to Phe (Wang et al., in press), and putative GSL products were induced by spraying leaves with 10 mM CuCl 2 ( aq.) ( Pedras et al., 2015). Hence, we decided to search for proposed aliphatic GSLs after various kinds of induction and from normal growth conditions. Specifically, we re-investigated both ecotypes of B. vulgaris for 2-propenylGSL (Pren, 107) and related aliphatic GSLs by ion trap HPLC-MS inspection for trace peaks, experimental testing of recovery of added GSLs, and investigation of plants challenged by herbivory and CuCl 2. We also investigated the morphologically deviating B. vulgaris ssp. vulgaris .
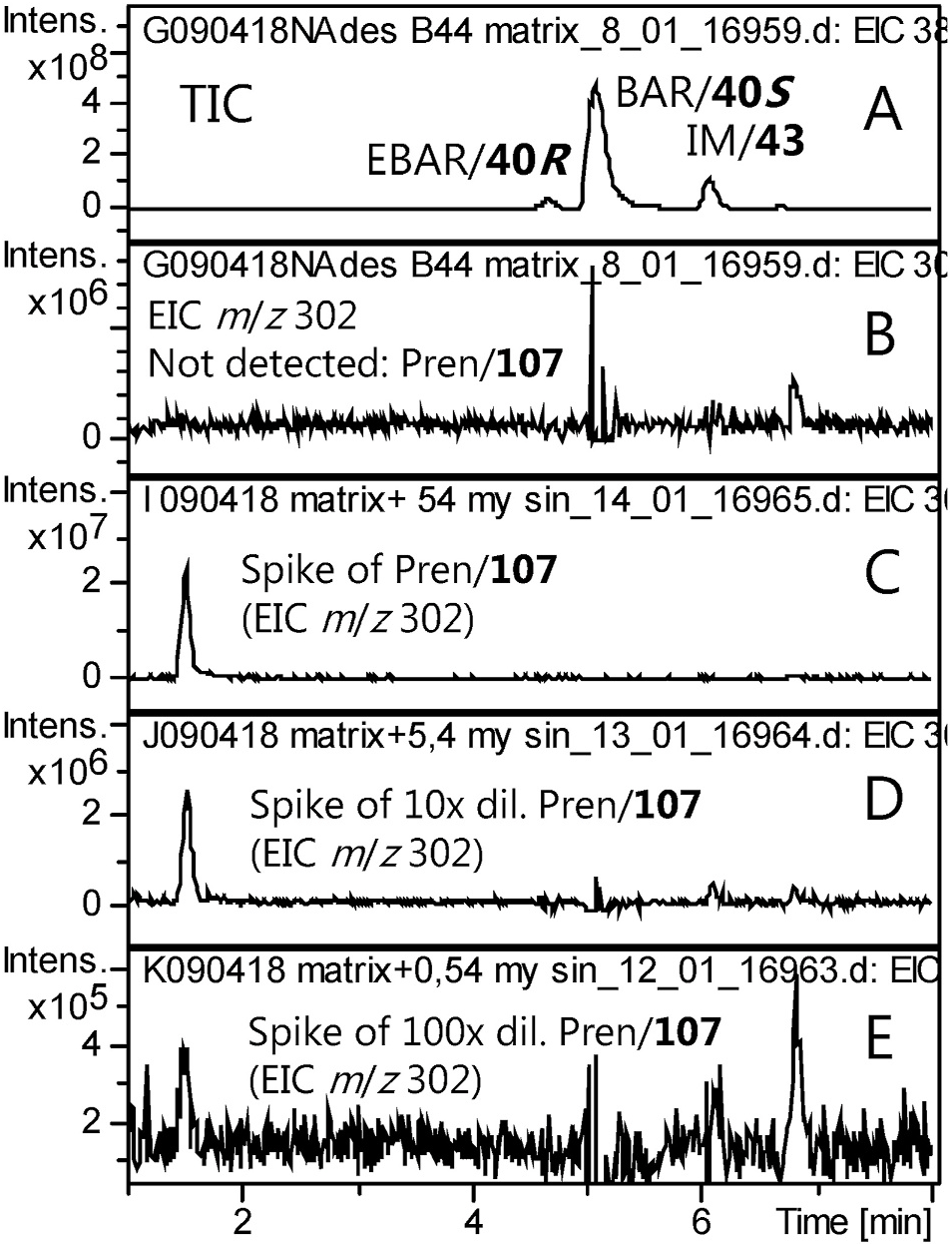
The GSL precursor of 2-propenyl ITC, Pren, was not detectable in B. vulgaris ( Fig. 6B View Fig ), in agreement with previous investigations of a range of P-type and G-type accessions from a large geographical range or harvested at different seasons (Agerbirk et al., 2015 and references cited therein). Experimental addition of (intact) Pren to crude extracts followed by desulfation confirmed linear detection of this GSL down to trace levels ( Fig. 6C–E View Fig ). The limit of detection was estimated as 0.1 μmol/g dry wt. at normal sample concentration ( Fig. 6E View Fig ) and 10-fold lower when using concentrated samples or high injection volumes. In the latter case, however, linearity of dominating GSLs suffered. The morphologically deviating ssp. vulgaris (in this report represented by accession B59), characterized by erect (vertical) rather than arcuate siliques, had a GSL profile indistinguishable from that of G-type plants (results not shown), in agreement with a previous report including ssp. vulgaris ( Agerbirk et al., 2003) .
We subjected P and G-type B. vulgaris to herbivory by larvae of the butterfly Pieris brassicae L. (3 days or 7 days), or by P. xylostella larvae (4 days), to spraying with 10 mM CuCl 2 (harvest after 4 days), and a parallel control treatment. These treatments were followed by GSL analysis using HPLC-UV for observing the general GSL profile and by ion trap HPLC-MS with injection of quite concentrated samples for a search for induced GSLs. The general leaf GSL profile was as expected for the type in both control and challenged plants, and we observed no candidate induced GSL peak. Some rather drastic treatments resulted in lower mean GSL levels, such as herbivory by the large P. brassicae for 7 days, after which time much of the leaves had been eaten ( Fig. 7 View Fig ). The lower GSL levels in remaining leaves may well be due to preferential ingestion of GSL rich plant parts of these GSL-stimulated larvae. We carefully searched for but did not detect Pren in any HPLC-MS sample in extracted ion chromatograms. Likewise, we systematically searched for but did not detect 1mEt, 1mPr, 2mPr, 2mBu, 2h2mBu, 3mPe, 2h3mPe, 3hmPe, 4mSb, 4mSOb, Buen, 8mSOo or BZ, selected from prominent GSLs in other tribe Cardamineae members and from GSLs produced by combination of A. thaliana and B. vulgaris biosynthetic enzymes after heterologous expression in tobacco (Wang et al., in press). Neither did visual inspection reveal any other new GSL for the species. However, we did detect trace levels of the known minor constituents 4mIM and PE, included in each search as a positive control.
During the final editing of the present work, a claim of ITCs corresponding to 3mSOp ( 73) and 4mSObuen ( 63) in P. xylostella -induced B. vulgaris was published ( Hussain et al., 2020). The claim, lacking actual evidense for the identifications as well as levels calculated on a plant weight basis, was based on analysis of G-type B. vulgaris of the Hedeland accession also used in our induction experiments, and no other isothiocyanate (such as the expected phenethyl ITC corresponding to PE) was reported ( Hussain et al., 2020). Hence, we searched three desulfoGSL analysis files of P. xylostella -induced B. vulgaris , including both the Hedeland and Suserup accessions, for any signs of d 63 or d 73. We also searched for d 95 from the expected biosynthetic precursor 95. No trace of any of these dGSLs was detected, although we are able to detect them in general ( Table 2 View Table 2 ). We also searched non-induced control samples with the same negative result. We notice that one of the claimed ITCs (from 73) would be isobaric with the expected ITC from PE ( 105), and that the corresponding dGSL (d 105) was detected as a minor constituent in several of our P. xylostella induced and control samples.
In conclusion, there are no reliable reports of aliphatic GSLs in the genus Barbarea . In contrast, extensive scrutiny has failed to reveal aliphatic GSLs above an estimated limit of detection of 0.1–0.01 μmol/g dry wt.
2.3. Phylogenetic analysis of Barbarea vulgaris View in CoL View at ENA accessions
In order to better understand whether the tested types and subspecies of B. vulgaris were representative for the entire species, we sequenced the nuclear ribosomal DNA internal transcribed spacer (ITS) region of a range of our accessions. For the P-type, we included our Danish “typeaccession” (B4) as well as three accessions representing major parts of northern Russia (Supplementary Table S1), and contrasted with two different morphological types of plants with glabrous leaves: the G-type (Danish “ type accession” B44) of ssp. arcuata (characterized by arcuate siliques) and a newly collected accession (B59) with erect (vertical) siliques traditionally named ssp. vulgaris by Danish botanists ( Hartvig et al., 2015).
The sequence variation of our accessions supported distinction of P- and G-types, with only two sequence types detected in the six sequenced accessions. The G-type as well as accession B59 of the morphological type traditionally known as ssp. vulgaris shared the same ITS sequence, while all four sequenced P-type accession differed from the G-type sequence at eight nucleotide positions ( Table 8 View Table 8 ) but where identical to each other.
This ITS sequence variation was subsequently compared with all B. vulgaris ITS sequences available from GenBank by mid-2019, most of them from Lange et al. (in review), resulting in a slightly more complex pattern. In this and the following discussion, we accepted the original sequence-contributors’ choice of intraspecific nomenclature. Currently, no intraspecific taxa are generally accepted and the intraspecific nomenclature of B. vulgaris is in need of revision (Lange et al., in review). The names represented with the GenBank material were: G-type, P-type and two traditionally recognized subspecies with more or less erect siliques; ssp. vulgaris and ssp. rivularis ( Hegi, 1958) .
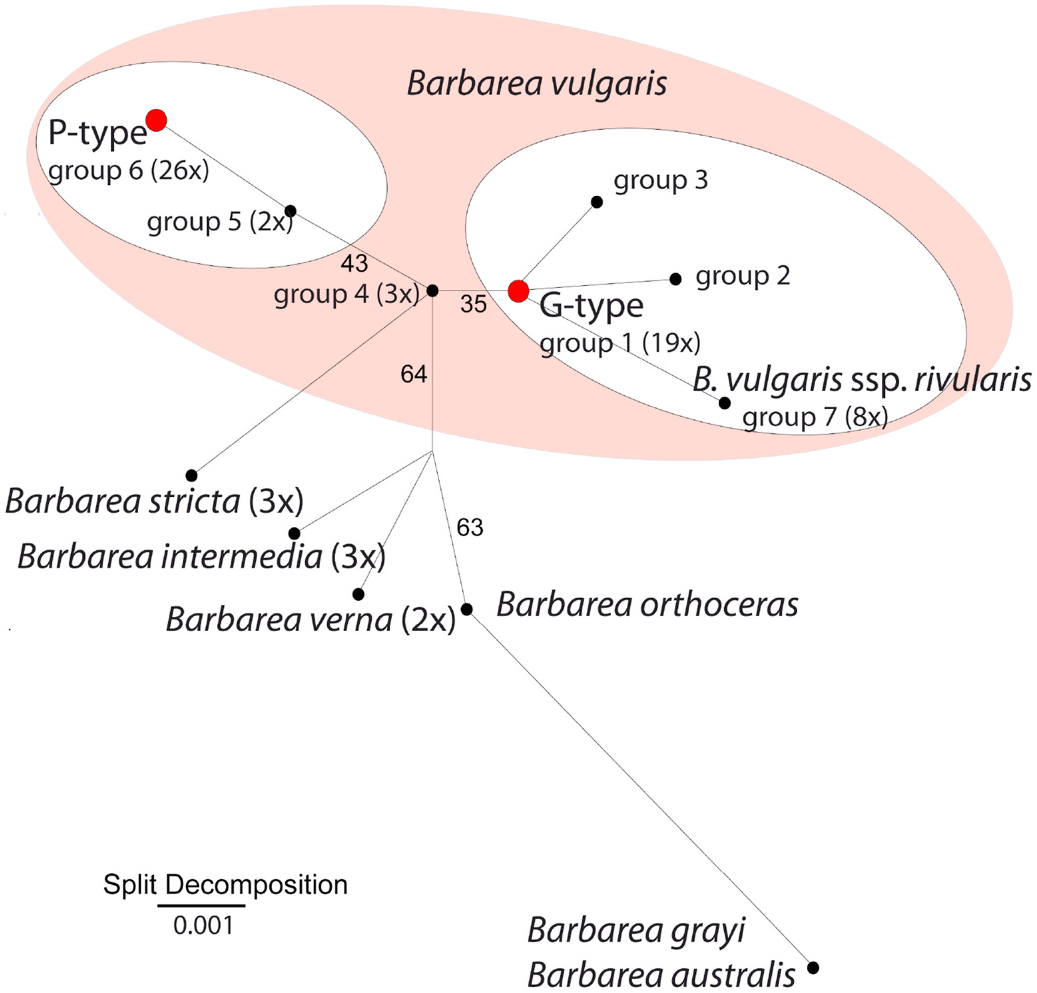
In total, seven ITS sequence variants were represented in GenBank (Suppl. Table S1, Table 8 View Table 8 ). The majority of these sequences likewise grouped with either our G-type or P-types in agreement with their respective type identities. One group (Group 4) was intermediary of the P-type and G-type sequences, and was tentatively interpreted as a hybrid of the G- and P-types. Indeed, one of them had been deposited as a hybrid sequence. Another group (group 7) was dominated by sequences deposited as ssp. rivularis and ssp. vulgaris and was more similar to the Gtype than to the P-type ( Table 8 View Table 8 ), for simplicity this group is named the rivularis group here, although the name may be temporary.
A few GenBank sequences did not agree with the group designations used here. Three sequences attributed to PxG hybrids clustered with the G-types in sequence group 1, as did two sequences attributed to ssp. vulgaris , including our accession B59. Likewise, one sequence attributed to G-type and two sequences attributed to hybrids grouped with the Ptypes in sequence group 6. Three other sequence groups (Groups 2, 3 and 5) represented sequences deposited as G-type, GxP hybrid or undetermined. These groups were represented by few deposited sequences.
The parsimony network analysis ( Fig. 8 View Fig ) suggested two main clusters of B. vulgaris , the one including the P-type and the small group 5 and the other including most of the remaining groups (G-type, rivularis and the small sequence groups 2 and 3). Group 4, tentatively interpreted as hybrid of P- and G-types, was intermediate. A separation of B. vulgaris into two gene pools, correlated with G-type and P-type, has also been shown earlier using different genetic marker systems ( Christensen et al., 2014; Toneatto et al., 2012).
In conclusion, the investigated G-type and P-type of B. vulgaris are rather representative for the known genetic variation of the species, which has mainly been investigated in the European part of its main original distribution area ( Christensen et al., 2014; Toneatto et al., 2010, 2012). Hence, lack of aliphatic GSLs may be a general property of the species (and genus). However, other sequence groups were found, with the “ rivularis ” group most deviating. These groups may represent other chemotypes as well. Hence, lack of aliphatic GSLs in the genus (Section 2.2.7.) can of course only be strictly concluded for the accessions investigated, not in general for the species and genus.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |