Ctenotus pantherinus (Peters, 1866)
|
publication ID |
https://doi.org/ 10.1093/zoolinnean/zlac076 |
|
publication LSID |
lsid:zoobank.org:pub:D6915151-59FA-4CE2-A059-E3FDE194BA3D |
|
DOI |
https://doi.org/10.5281/zenodo.7696074 |
|
persistent identifier |
https://treatment.plazi.org/id/03FFAB1A-0743-CA1C-5647-6A59FAF7F88D |
|
treatment provided by |
Plazi |
|
scientific name |
Ctenotus pantherinus |
| status |
|
SUPPORT FOR C. PANTHERINUS SUBSPECIES
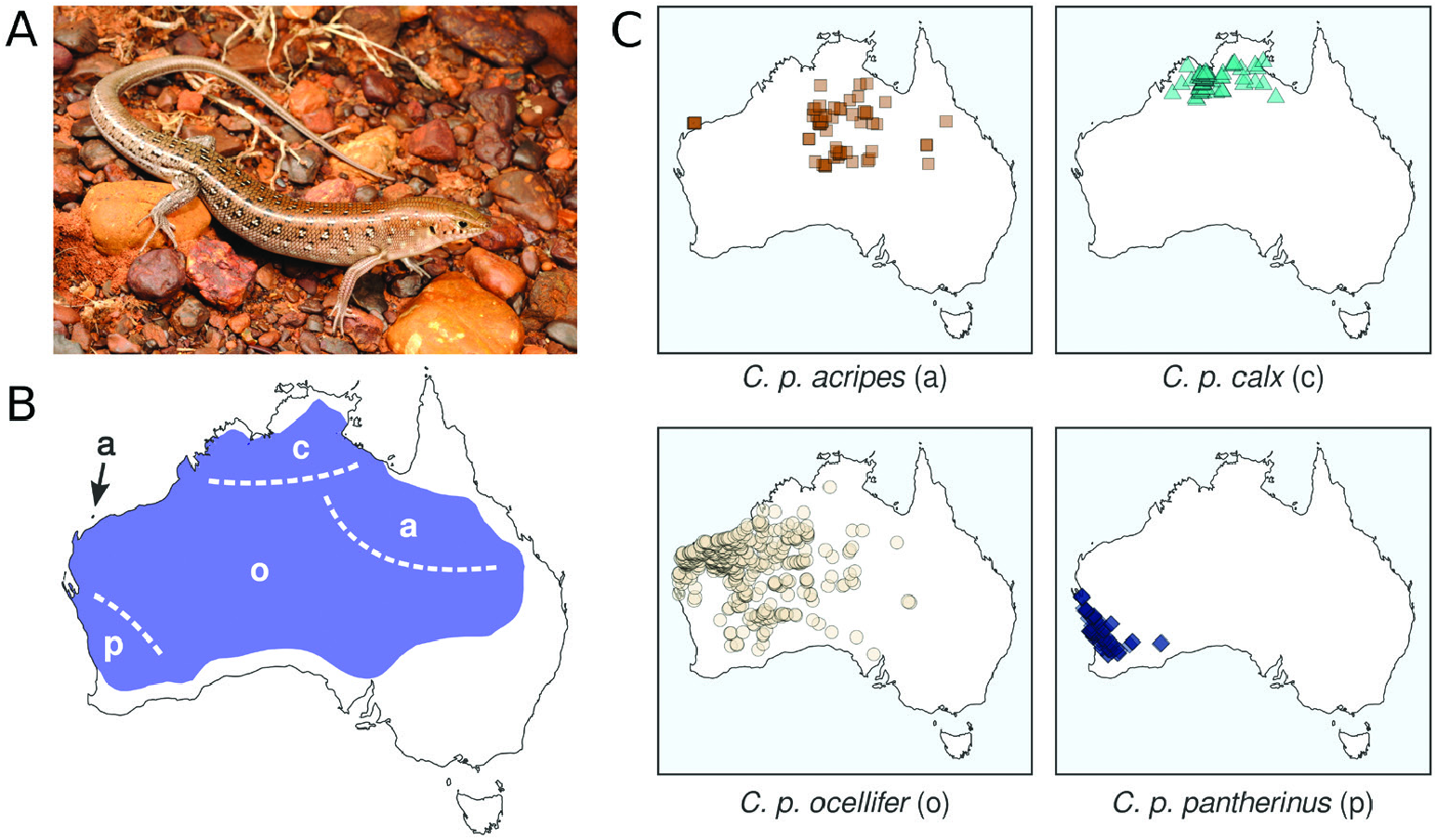
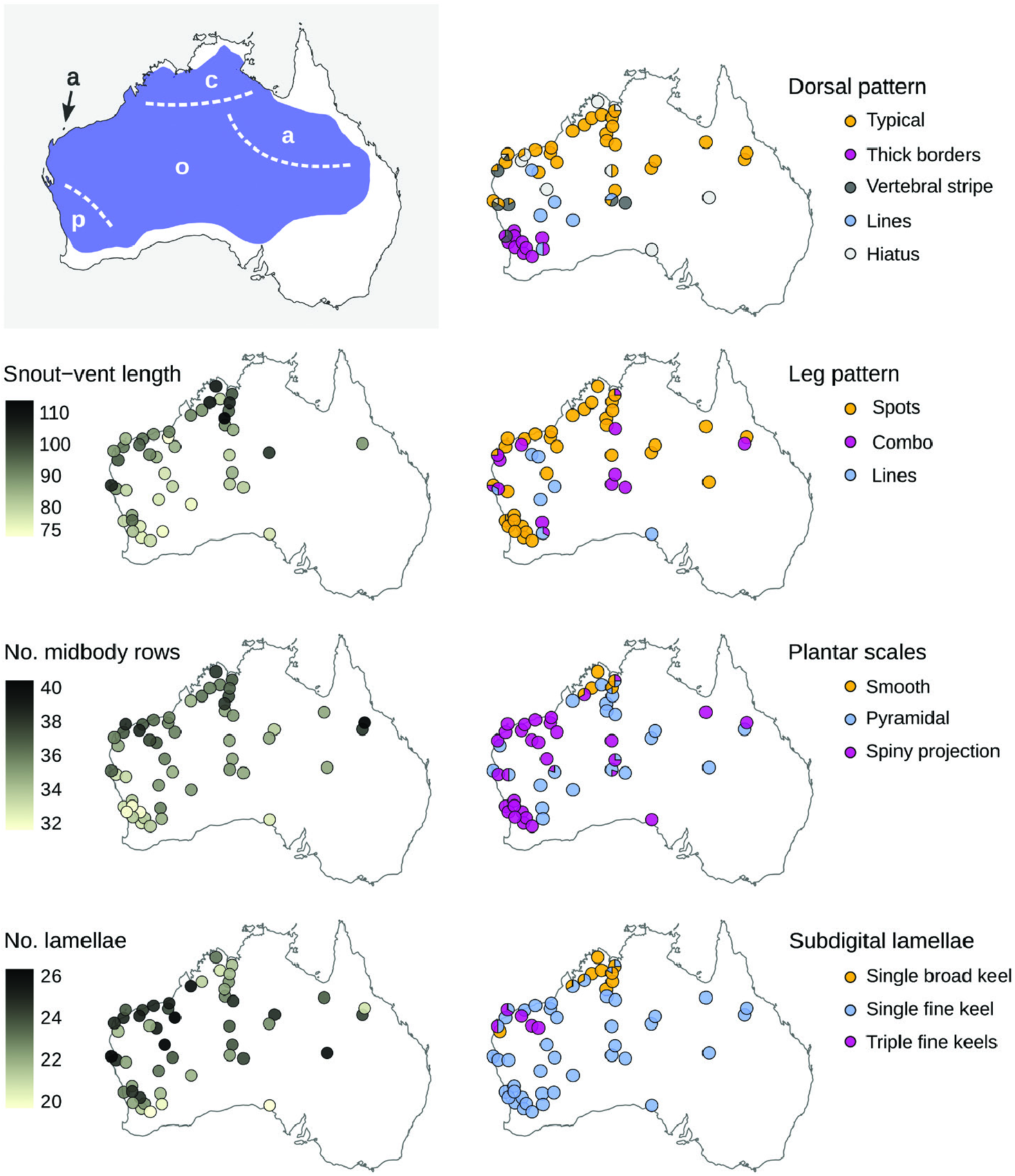
Our morphological examinations confirmed that coloration and scalation characters broadly used in Ctenotus taxonomy varies across the range of C. pantherinus . Some character states were more common in certain regions, including broadly keeled digits, lower midbody scale counts, larger average adult sizes and dorsal spots with thick outlines. This variation was consistent, in part, with the presumed distributions of certain subspecies, namely C. p. calx and C. p. pantherinus . Nevertheless, many individuals in a given region deviated from the most frequent phenotype and corresponding phylogenetic lineage, whereas character combinations presumed as diagnostic also occurred outside the recognized subspecies ranges. Such regional variation appears to have been overlooked or unreported in the subspecies descriptions, potentially owing to limited sampling. In the case of the continuous characters, spatial variation often appeared clinal. Moreover, despite broad geographical trends when we examined certain characters in isolation, the morphological distinctiveness of regional populations faded when multiple characters were considered jointly. Concentrating on single characters and overlooking their intrasite and intraregion variation appears to have overestimated the phenotypic coherence and distinctiveness of the subspecies of C. pantherinus .
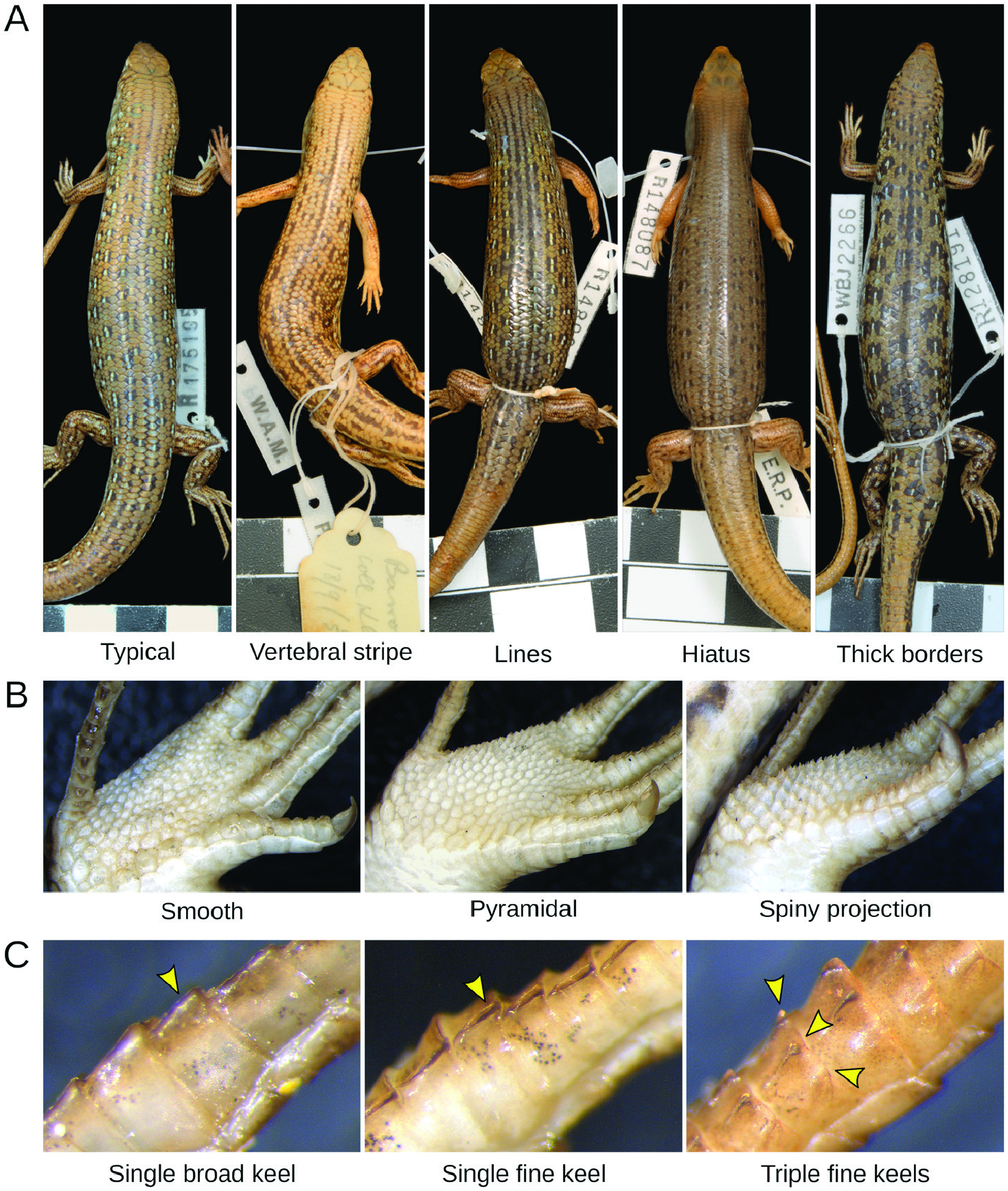
Other patterns of morphological variation did not conform to subspecies definitions. We found some character states to be infrequent in regions where these states were once thought to predominate, whereas others occurred more extensively than previously reported. Most of the character variation we encountered was continuous (e.g. degree of spininess of the plantar scales and coloration traits; Fig. 2 View Figure 2 ), making it difficult to score these characters objectively. This difficulty might explain the pattern of conflicting voucher assignments among collectors and museum staff ( Fig. 1 View Figure 1 ) and our partial failure to recover previously reported spatial patterns ( Fig. 3 View Figure 3 ; Table 1 View Table 1 ). Remarkably, some of the subspecies did not appear morphologically distinct at all (C. p. acripes and C. p. ocellifer). These findings might stem from broader specimen collections now available relative to when the subspecies were described. For instance, <100 specimens of C. pantherinus were housed at the Western Australian Museum at the time C. p. calx was described based on nine of them. Likewise, ~ 200 specimens of C. pantherinus were available in the same museum when C. p. acripes was described, with that description incorporating 28 specimens from a single site ( Barrow Island ). Today, this museum houses> 1300 C. pantherinus specimens. This increased sampling has revealed variation that contradicts the presumed coherence and distinctiveness of subspecies .
Similar to the morphological patterns, the correspondence of genetic clades to currently recognized subspecies was weak and inconsistent. Samples from the presumed range of a subspecies were partitioned into multiple clades, particularly in the nuclear analysis. Conversely, clades that corresponded roughly to a subspecies included samples from another presumed distribution of a subspecies. Notably, the mitochondrial analysis grouped two to three subspecies together and inferred all subspecies as paraphyletic. This finding appears unexpected, because shorter coalescent times in mitochondrial markers (relative to nuclear DNA) make them well suited to the identification of shallow divergences, such as those seen within species ( Palumbi et al., 2001). Instead, the results are consistent with mitochondrial haplotype sharing among populations of the same species. Nuclear estimates of ancestry coefficients and widespread mitonuclear discordance provide further support for a pattern of broad genetic admixture and introgression, as reported in other species (e.g. Toews & Brelsford, 2012; Pereira et al., 2016). Therefore, although the genetic data support the presence of multiple incipient lineages within C. pantherinus , these lineages do not appear to be evolving independently, providing no support for their designation as species-level taxa.
CHALLENGES TO THE TAXONOMIC CATEGORIZATION OF EARLY DIVERGING LINEAGES
Genetic admixture and introgression between major genetic groups in C. pantherinus arguably provide evidence of incomplete evolutionary separation, consistent with certain conceptualizations of subspecies ( Frost & Hillis, 1990; Hillis, 2020; de Queiroz, 2020, 2021). Genetic exchange across incipient lineages might explain why many sampled localities were polymorphic and a pattern of widespread character state sharing across regions. In the presence of gene flow, populations can develop misaligned phenotypic and neutral genetic transitions in space ( Lipshutz et al., 2019). Therefore, individuals with conflicting phenotypic and genetic patterns, as seen in C. pantherinus , might be typical of the early stages of lineage divergence ( Zamudio et al., 2016).
Facing such instances of regional variation, some authors have proposed the recognition of a subspecies when an arbitrary proportion (e.g. 75%) of the individuals in a region exhibit a given trait ( Amadon, 1949; Patten & Unitt, 2002). This proposal illustrates that, if we decide to categorize incipient lineages in taxonomy, they can be described at best in terms of the most frequent phenotypes and clade memberships. Under such a scheme, an unknown, variable and potentially large proportion of individuals will be misclassified or classified ambiguously. We might expect the spatial limits of subspecies ranges to be unclear, not only because of phenotypic or genetic clines but, perhaps primarily, owing to fallible morphological and genetic diagnoses. As suggested by empirical analyses of mammals (e.g. Patton & Conroy, 2017), birds (e.g. Patten & Unitt, 2002), butterflies (reviewed by Braby et al., 2012) and our analyses of C. pantherinus , the evolutionary processes that characterize early lineage divergence might preclude the unequivocal assignment of many individuals to taxa. It seems likely that researchers will continue to disagree on whether this limitation undermines or justifies the utility of subspecies in taxonomic practice ( Patton & Conroy, 2017).
A recent debate on subspecies has revolved around redefining this category to indicate incompletely separated population lineages ( Hillis, 2020; de Queiroz, 2020, 2021). The case of C. pantherinus suggests that, despite the conceptual appeal of this redefined subspecies concept, it might be unclear how to use it to guide taxonomic practice. It can be challenging to determine whether a pair of population lineages is separated completely or incompletely. For instance, closely related populations frequently show varying degrees of genetic divergence and admixture ( Singhal & Moritz, 2013; Dufresnes et al., 2015). This pattern raises the question of what degree of genetic divergence or reduction in gene flow might warrant the recognition of species or subspecies ( Padial & De la Riva, 2020). To circumvent this issue, we might consider as conspecific those population lineages showing any level of incomplete separation. We might then propose subspecies to indicate identifiable genetic and morphological subgroups. However, many divergent lineages experience rampant genetic introgression; arguably, the most direct indication of incomplete lineage separation. Often, these lineages belong to distant (e.g. genus-level) clades and differ starkly in morphology, ecology and behaviour (e.g. hybridizing ducks or canids) ( Johnsgard, 1960; Monzón et al., 2014). Despite their apparent incomplete separation, such lineages would hardly be considered conspecific. These examples illustrate some of the challenges in translating conceptual definitions of taxonomic categories into empirical taxon delimitation. Such challenges apply to both subspecies and species ( de Queiroz, 2007).
TRADITIONAL SUBSPECIES ARE DIFFICULT TO TEST AND FALSIFY
This study also highlights another peculiarity of subspecies: a historical asymmetry, whereby subspecies proposed in the past are hard to test and falsify ( Burbrink et al., 2022), contrasting with an apparent hesitation from the taxonomists of today to propose new subspecies. In reptiles, for instance, subspecies descriptions peaked around the 1960s but declined sharply thereafter despite, or maybe because of, a rapid increase in specimen collection towards the end of the 20 th century ( Uetz & Stylianou, 2018). Subspecies proposed decades ago can be difficult to falsify and discard owing to typically vague morphological definitions and deference to the opinions of previous workers about population distinctiveness. Given that subspecies are nested in a developmentally and ecologically constrained species, the phenotypic differences invoked to define subspecies are necessarily subtle. Furthermore, as in C. pantherinus , subspecies were often described focusing on one or a few characters from relatively little material, thereby sampling gaps might have exacerbated the perception of distinctiveness ( Braby et al., 2012). Given that trait variation can be seen even when incompletely characterized, and owing to broad acceptance of partly speculative geographical ranges (e.g. Fig. 1 View Figure 1 ), even vaguely defined subspecies continue to be recognized in taxonomic treatments and field guides. In contrast, present-day taxonomists appear unlikely to propose subspecies based on evidentiary standards typical in the 1960s–1980s. As a case in point, a recent study on Australian frill-necked lizards ( Chlamydosaurus kingii Gray, 1825 ) described clinal variation in frill colour over the distribution of this species ( Pepper et al., 2017). However, no subspecific taxa have been proposed to accommodate this variation, despite the presence of concomitant (albeit shallow) genetic differentiation. In contrast, recent evidence of limited or inconsistent distinctiveness in phenotype and genotype does not appear to bear on the rejection of many historically proposed subspecies ( Zink, 2004).
Contrasting with the view that subspecies must correspond to lineages, some researchers advocate for using subspecies to denote groups of phenotypically similar populations regardless of evolutionary relationships ( Patton & Conroy, 2017). This perspective is at odds with principles of scientific thought that trace back to Darwin, whereby taxa at all levels of biological classification should reflect phylogenetic relationships ( Darwin, 1859; de Queiroz & Gauthier, 1992). This criterion also applies to the species category, because broadly applied concepts define species as phylogenetic lineages or predict that they will become lineages through sustained reproductive isolation ( Hennig, 1966; Dobzhansky, 1971; Cracraft, 1987; de Queiroz, 1998; Harrison & Larson, 2014). In this regard, it is worth noting that the nature of species as lineages is unaffected by the inference of paraphyly in gene genealogies (e.g. from incomplete lineage sorting or introgression; Padial & De la Riva, 2020). In contrast, a strictly morphological subspecies concept disregards the otherwise universal criterion of phylogeny, and thus potential incongruences between phylogeny and phenotype ( Burbrink et al., 2000). Under this concept, ‘subspecies’ might evoke similarly named categories, such as subgenus or subfamily, but is the only one not required to denote a clade. As such, ‘subspecies’ can be a misleading term because it is considered a taxonomic category but lacks the defining property of all other taxonomic categories.
Additionally, by overlooking the evolutionary coherence of populations, morphological subspecies definitions are unfalsifiable. As illustrated by C. pantherinus , such definitions require only that individuals from distinct locations tend to differ in a given trait, even if trait variation within and across locations hampers subspecies diagnosability ( Patten & Unitt, 2002; Braby et al., 2012). Thus, limited internal coherence does not appear to challenge morphologically defined subspecies, contrasting with taxonomic groupings based on evolutionary relationships. Nevertheless, some authors have argued that even morphologically diagnosable subspecies are not biologically meaningful unless their defining characters reflect evolutionary separation ( Mayr, 1963; Reydon & Kunz, 2021). Otherwise, the characters used to identify subspecies are essentially arbitrary, and infinite partitions could be proposed within any species ( Mayr, 1963; Wilson & Brown, 1953). Given the long-lasting contentions on how to designate subspecies, it might be clearer to simply annotate phenotypic variation patterns across species ranges as relevant (e.g. Owen, 1963a, b). This approach does not artificially impose discrete taxonomic structures on clinal or other continuous patterns of variation ( Wilson & Brown, 1953; Mayr, 1963; Owen, 1963b).
Finally, some authors advocate using geographical range as a ‘diagnostic character’ of subspecies ( Patton & Conroy, 2017). Although this often allows fieldworkers to assign taxonomic labels to specimens more easily, this solution is inherently circular and leads, in a similar manner, to taxonomic entities that cannot be falsified. Moreover, as illustrated by C. pantherinus , reliance on geographical location for subspecies assignment can result in groupings of individuals that lack morphological and genetic coherence. Other studies have found that subspecies defined primarily based on geographical ranges are not phylogenetically divergent from those in neighbouring regions, as is the case of insular populations of primates in eastern Africa ( Penna et al., 2022).
IMPLICATIONS FOR THE INFRASPECIFIC TAXONOMY OF C. PANTHERINUS
Patterns of genetic and morphological variation appear to contradict the presence of independently evolving lineages and thus unrecognized species diversity within C. pantherinus . Additionally, our results provide weak to no support for the currently recognized subspecies and their presumed distributions. The data at hand challenge the distinction of C. p. acripes (Barrow Island and north-eastern Australia) from C. p. ocellifer (arid zone) owing to extensive paraphyly and morphological overlap. Nuclear (but not mitochondrial) markers and a couple of morphological characters appear consistent only in part with C. p. pantherinus (south-west) and C. p. calx (north). However, intrasite polymorphism is prevalent, and morphological characters proposed as diagnostic broadly occur outside the purported range of a subspecies. In the face of these patterns, we struggled to assign individuals to subspecies based on morphological characters alone. Moreover, we failed to match the genetic lineages and spatial character transitions we have identified with subspecies ranges as currently understood.
Arguably, the four traditional subspecies capture certain aspects of phenotypic or genetic variation across the range of C. pantherinus . Nonetheless, it seems unlikely that a modern taxonomist with access to the data presented here would converge on the proposed subspecies definitions. Even if particular coloration and scalation traits show some degree of geographical structure, no partition of characters is diagnostic of regional populations, owing to local polymorphisms and shared character states across regions. Moreover, genetic data do not support a partition of monophyletic units corresponding to phenotypes. Taken together, these results suggest that it is appropriate to synonymize Lygosoma ocelliferum (= Ctenotus pantherinus ocellifer ), Ctenotus pantherinus calx and Ctenotus pantherinus acripes with Ctenotus pantherinus , and we do so formally here. Should future evidence support the recognition of new or redefined taxa (including species), the northern and south-western names are available. However, it seems unlikely that the name C. p. acripes might be found to correspond to a distinctive unit. At this time, recognizing C. pantherinus as a wide-ranging species devoid of subspecific taxa appears best to convey the evolutionary history of the species and often spatially idiosyncratic patterns of phenotypic variation. We believe this fundamentally conservative arrangement is preferable to recognizing subspecies whose unclear boundaries, distributions and morphological diagnoses have baffled many biologists working in the field and in natural history collections ( Fig. 1 View Figure 1 ).
Patterns of partly decoupled morphological and genetic transitions across the range of widespread species, as seen in C. pantherinus , provide opportunities to investigate the factors behind lineage and trait divergence ( Lipshutz et al., 2019). To identify the incipient lineages involved properly, presentday researchers will need to reassess many taxa that were proposed using small datasets and under fundamentally different views about the nature of species and other taxonomic categories ( Zink, 2004; Padial & De la Riva, 2020; de Queiroz, 2020).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.