Eulaemus, Girard, 1857
|
publication ID |
https://doi.org/10.1111/zoj.12231 |
|
persistent identifier |
https://treatment.plazi.org/id/03E02B2A-FFC4-4A3A-D6AA-C306FE60FF04 |
|
treatment provided by |
Felipe |
|
scientific name |
Eulaemus |
| status |
|
EULAEMUS PHYLOGENY
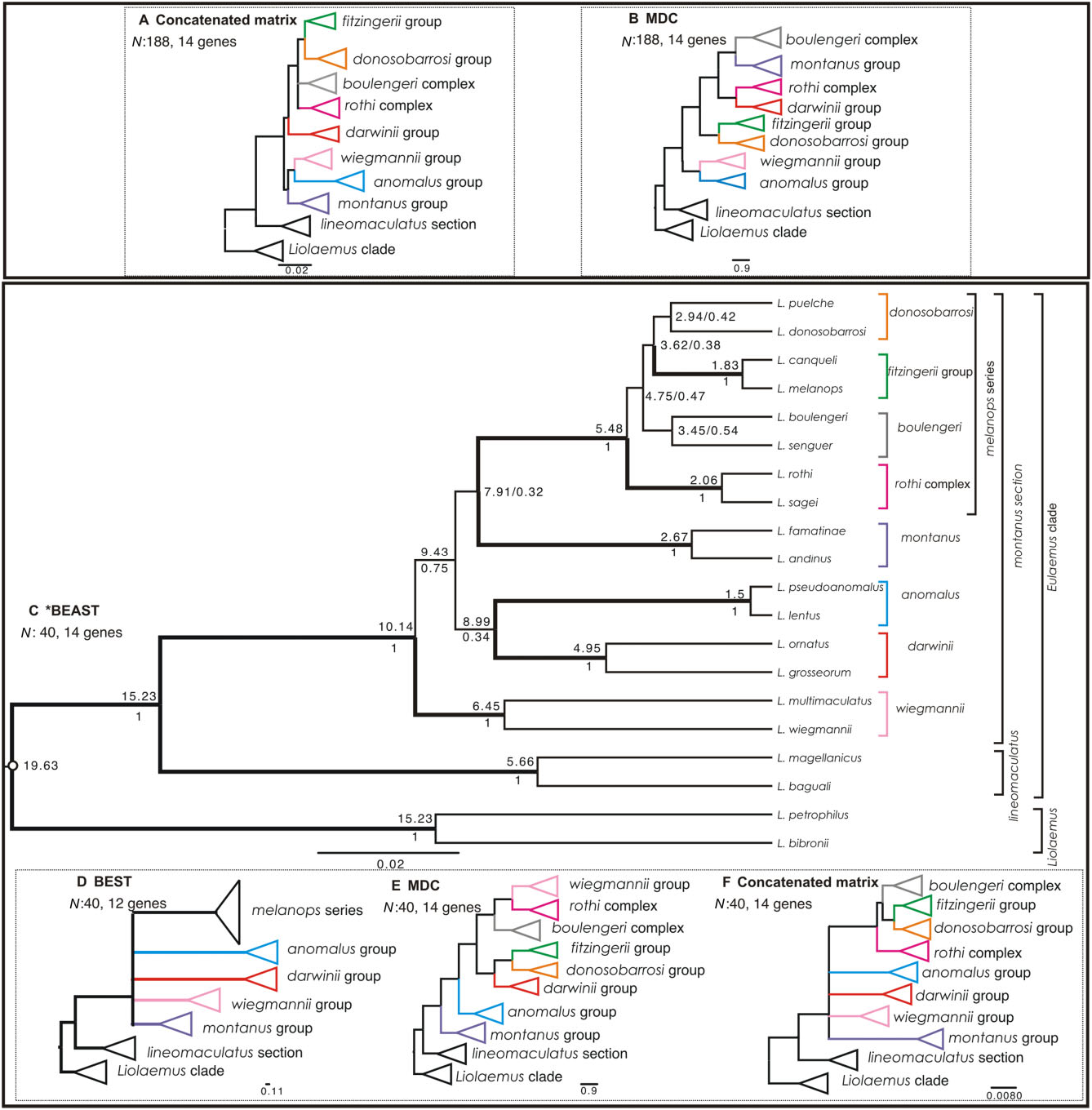
Here we used four different methods representing different conceptual approaches to reconstruct phylogenies of the subgenus Eulaemus . First, we used a complete matrix of 188 terminals and 14 loci (MDC and concatenated analyses), and then selected representatives of the main clades for which we implemented two Bayesian species tree methods (*BEAST and BEST). All of our analyses strongly support the lineomaculatus section as the sister clade of the montanus section ( Fig. 3 View Figure 3 A-F). We also recovered identical main clades within the montanus section with all four methods, with the exception of the paraphyletic melanops series’ groups in the MDC analysis ( Fig. 3E View Figure 3 ), but these were recovered as monophyletic in the MDC and the concatenated analysis for the full matrix ( Fig. 3A, B View Figure 3 ). However, we did not fully resolve rela- tionships amongst these main clades owing to short, weakly supported internodes, and the same was true for relationships amongst the main clades within the montanus section ( Fig. 3A, C View Figure 3 ).
Previously published phylogenetic studies of the genus Liolaemus show extensive topological incongruence (e.g. Etheridge, 1995, 2000; Schulte et al., 2000; Espinoza, Wiens & Tracy, 2004; Avila et al., 2006; Abdala, 2007; Fontanella et al., 2012), with few shared strongly supported hypotheses; thus, no consensus has been reached. In this most inclusive study to date, we did not recover a strongly supported and fully resolved phylogeny, and we represent the uncertain relationships as polytomies. Although these polytomies may be re- solved by adding more data, they may also indicate rapid radiations amongst some clades. However, before arriving at this conclusion, we need to consider that poor branch support can also be the result of: (1) insufficiently informative data; (2) data sets that strongly conflict with one another; (3) inappropriate phylogenetic methods and substitution models; or (4) insufficient data to resolve short branches ( Whitfield & Lockhart, 2007).
To address point 1, we explored the informativeness of our data set using likelihood-mapping as well as saturation tests and phylogenetic signal index calculations. These analyses showed that all of the 13 loci included in our empirical analyses and simulations are phylogenetically informative, and although it was not possible to resolve the relationships amongst the main clades within the montanus section, our data are informative enough to resolve the oldest divergence (between the montanus and lineomaculatus sections), as well as providing high levels of resolution within each main clade (results not shown; see Olave et al., 2014). This suggests that our data set is sufficiently informative to resolve recent and ancient divergences, probably because we included loci with relatively high (12S and cyt-b), intermediate (KIF24, A12D, A1D, A4B, A9C), and low substitution rates (EXPH5, PRLR, SNCAIP, CMOS, DNAH3, MXRA5, PNN; see details in Table 2). In our experience with other lizard clades, some combination of these or similar loci is usually sufficient to recover well-resolved/wellsupported trees ( Benavides et al., 2009; Sinclair et al., 2010; Breitman et al., 2011; Camargo et al., 2012; Werneck et al., 2012). Further, Camargo et al. (2012) showed in simulation studies that the accuracy of Bayesian species tree methods is significantly higher when multiple loci of different mutation rates are used. Multiple samples per species are also necessary for successful estimation of species trees in *BEAST and although we had to reduce the number of samples per species, simulation studies show that even two samples per species are sufficient given enough loci ( Heled & Drummond, 2010).
To minimize the effects of points 2 (conflicts in data) and 3 (inappropriate methods), we used three recently developed approaches (*BEAST, BEST, and MDC) that accommodate discordance amongst gene trees to estimate species trees. Finally, to address point 4 (insufficient data) we included the largest molecular data set and the most dense species sampling effort (188 terminals, 14 loci) of any phylogenetic study of this genus.
After considering all of these likely causes of poor phylogenetic reconstruction and still not resolving some polytomies, we performed statistical tests of diversification hypotheses within the montanus section. We tested two models with one and two hard polytomies (with estimated divergence times of 10.14 and 5.48 Mya, respectively) in the broader context of five models based on different published topologies and a sixth alternative based on our *BEAST results (Fig. 1). We found some support for all these models ( Fig. 3 View Figure 3 ), but the strongest support favoured the ‘two hard polytomies’ model (nine loci), followed by the ‘one hard polytomy’ model (eight loci). The models based on the Schulte et al. (2000) hypothesis and our *BEAST analysis were supported by seven loci, and the Avila et al. (2006) topology was supported by six loci.
Hard polytomies are recognized by very short internodes for which by chance every descendent lineage has the same probability of receiving one allele ( McCracken & Sorenson, 2005). This implies that when multiple loci are analysed we would expect to find support for different gene tree topologies owing to a stochastic pattern of shared allele sorting amongst lineages. As the length of the internode increases, an increasing proportion of gene trees should become congruent with the species history.
Our tests suggest that two hard polytomies are the most plausible explanation for the history of this clade amongst the eight models evaluated. However, the difference between two and one hard polytomy models is only one locus, and we note that differences probably reflect the uncertainty of a real statistical difference between these results. As the ‘two hard polytomies’ hypothesis is the most strongly supported, and because we recovered the melanops series as monophyletic in most phylogenetic analyses ( Fig. 3A, C, D, F View Figure 3 ) with a longer average speciation time (7.91–5.48 = 2.33 Mya interval), we accept this model as the best working hypothesis. If rapid simultaneous radiations of lineages is the true history for this clade, then the incongruence amongst previously published studies is expected; all of these studies found some well-supported topological differences amongst the main clades regardless of the method or data set used.
Phylogenetic methods are designed to locate dichotomies in trees, and until recently none was appropriate to search for a shared MRCA amongst three or more lineages. Traditional concatenated analyses also tend to inflate nodal support for dichotomies that may not be real ( Belfiore, Liang & Moritz, 2008), but modelbased approaches now provide new analytical possibilities ( Knowles, 2009), and we designed such a test here to shed light on the evolutionary history of the Eulaemus clade. Our results suggest that the most plausible species tree for this clade includes two hard polytomies amongst lineages, and describes two events of rapid radiation of lineages in Eulaemus history. If true, then we predict that neither the inclusion of species not sampled here, nor the increase in the number of informative loci, will resolve these polytomies ( Delsuc, Brinkmann & Philippe, 2005; Rokas & Carroll, 2006; Whitfield & Lockhart, 2007).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.