Danio
|
publication ID |
https://doi.org/ 10.1046/j.1096-3642.2002.00014.x |
|
publication LSID |
lsid:zoobank.org:pub:3CCFE85A-2CCE-4A40-8AAA-229DCAFAA958 |
|
persistent identifier |
https://treatment.plazi.org/id/03DDFD6F-FFA7-1E29-914D-64E6FE89F988 |
|
treatment provided by |
Carolina |
|
scientific name |
Danio |
| status |
|
DANIO View in CoL View at ENA OSTEOLOGY
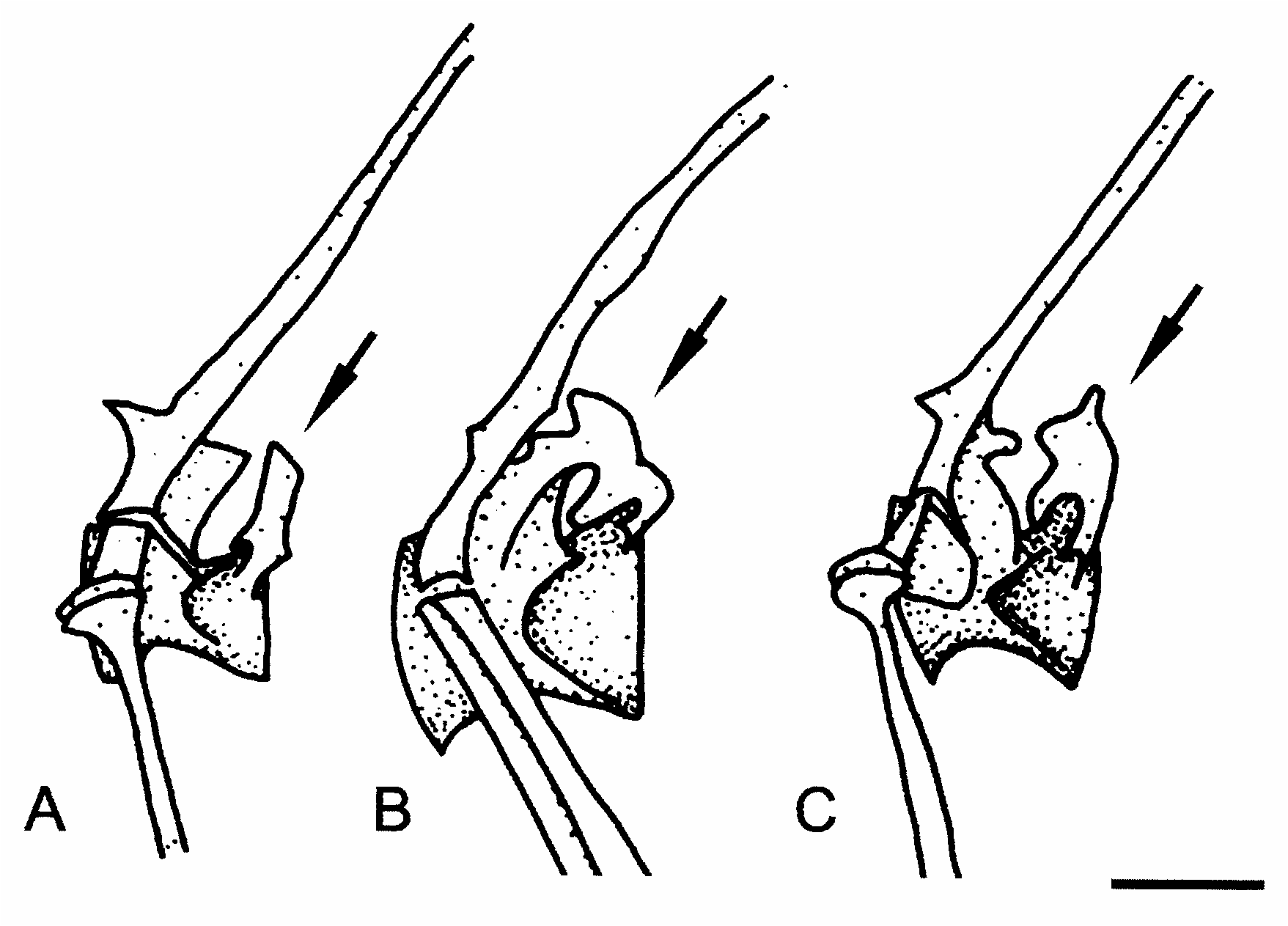
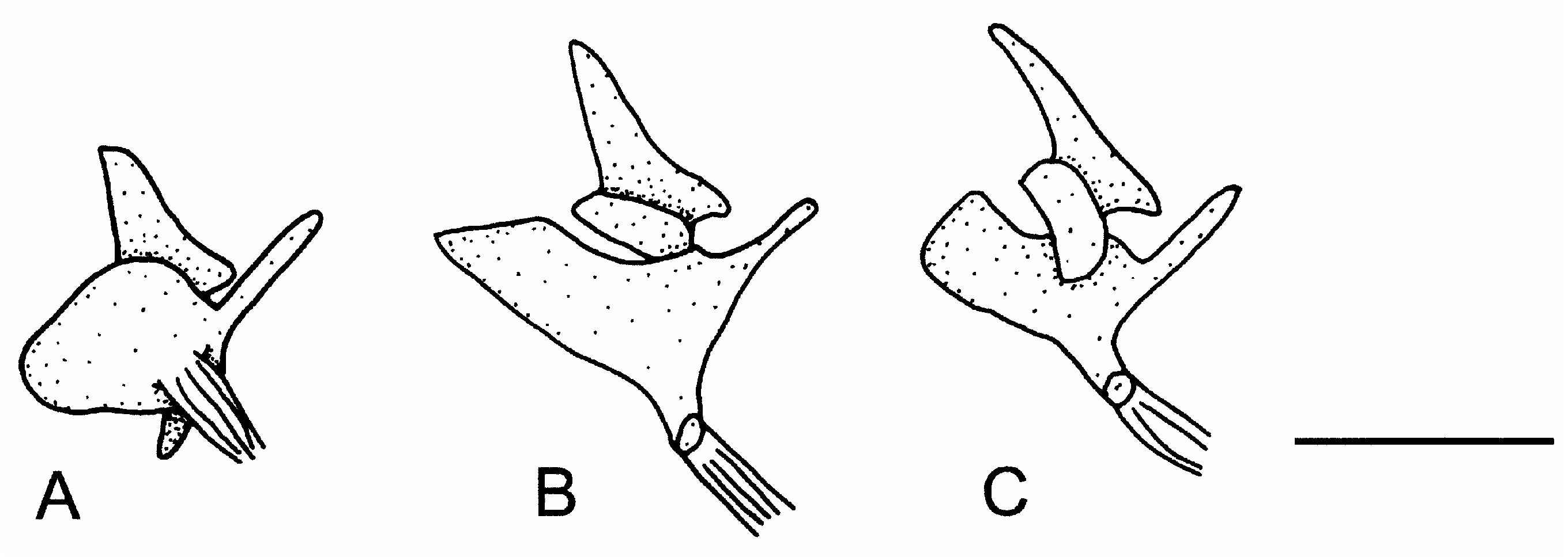
Osteological studies often rely on only a few specimens of cleared and stained material for each species of interest, making it difficult to observe intraspecific variation or ontogenetic changes in osteology. Furthermore, the emphasis in ontogenetic studies is generally on early ontogenetic stages. However, late ontogenetic changes have been observed in paddlefish ( Grande & Bemis, 1991) and some cyprinids ( Cavender & Coburn, 1992). The lack of attention paid to late ontogenetic stages has, in some cases, been the source of apparent conflicts in the literature (see Grande & Bemis, 1991). In older individuals of Danio we have also observed a higher frequency of growths and reductions. For example, the posterior processes of the fifth centrum in D. kerri seem to undergo changes with growth ( Fig. 10 View Figure 10 ). This is exemplified in one individual where this process has fused with the posterior flange of the neural arch ( Fig. 10b View Figure 10 ). The ligamentous attachment on the scaphium also seems to become elongated in older specimens of D. kerri , though this observation is based on only three to four individuals of each tentative age class ( Fig. 11 View Figure 11 ). Older individuals of D. kerri and D. browni also commonly exhibit a thinning or complete loss of bone at the articulation of the scaphium and claustrum ( Fig. 11b,c View Figure 11 ). Fusions of vertebrae two and three are common within Cypriniformes ( Fink & Fink, 1981, 1996; Cavender & Coburn, 1992) but the actual distribution of this character is uncertain ( Cavender & Coburn, 1992). Cavender & Coburn (1992), noting that a better understanding of this fusion’s ontogeny is needed, suggest that this fusion may occur earlier in ontogeny in the subfamily Cyprininae than in the subfamily Leuciscinae . In one older individual of D. browni there are notable calcifications between vertebral centra two and three while there are no fusions or calcifications in any other individuals of this species examined.
PHYLOGENETIC ANALYSIS
Morphological analysis
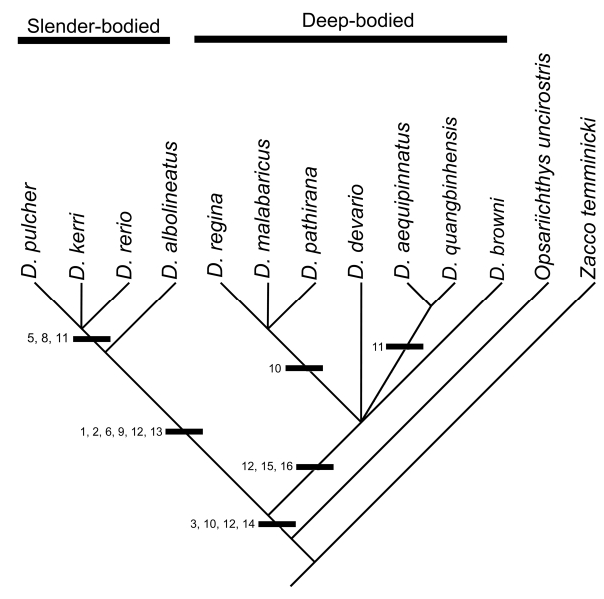
Study of the axial skeleton in 11 species of Danio yields 14 characters that are potentially useful in phylogenetic analysis. Two pigmentation characters were also incorporated from McClure (1998, 1999). Characters, as described in the preceding text, are summarized in Table 2. Table 3 gives the character matrix used for phylogenetic analysis. The consensus phylogeny derived from two equally-parsimonious tree is illustrated in Figure 12 View Figure 12 . According to our analysis, four morphological characters support the monophyly of Danio . These characters include: the lack of an anterior groove in the large supraneural (character 3), reduction of the ascending process of the intercalarium (character 10), having 38 or fewer vertebrae (character 12), and presence of a space between the epural and rudimentary neural arch of the compound centrum (character 14). Study of additional Danio , rasborin and cyprinid species is desirable, however, to confirm these characters. Our analysis also shows strong support for the basal division of Danio into deep-bodied and shallow-bodied subclades, though resolution within these subclades is low. The slenderbodied subclade is supported by six synapomorphies: having a total vertebrae count of 31–33 (character 12), loss of the sixth hypural (character 13), presence of a medial flange on the os suspensorium (character 1), blade-like fourth neural spine (character 2), lack of a distal ridge on the tripus (character 9), and a foreshortened large supraneural (character 6). Three characters support the deep-bodied subclade: having a total vertebrae count of 34–38 (character 12), reticulations in melanophore patterning (character 15), and presence of a postopercular spot (character 16).
Molecular analysis
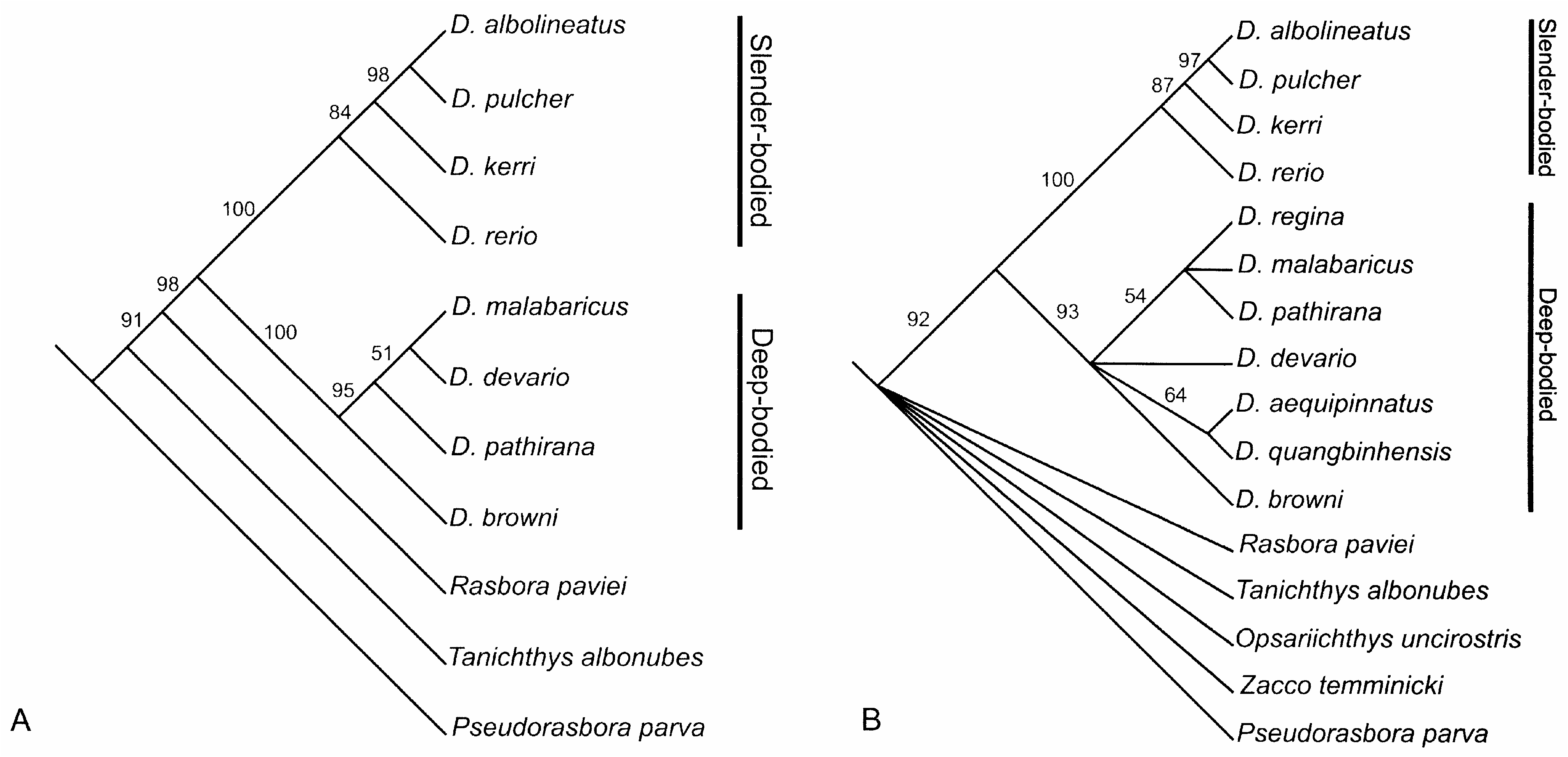
Combined sequences from five genes comprising 2189 total base pairs yields 246 potentially informative sites. Table 4 gives partitions of the total variable sites and informative sites by gene. Molecular data also strongly support the monophyly of Danio as well as the basal division into slender and deep-bodied subclades. The bootstrap consensus molecular phylogeny derived from two equally-parsimonious phylogenies is illustrated in Figure 13 View Figure 13 (a). The two equally- parsimonious trees differ only in the relationship of D. malabaricus to D. pathirana and D. devario . Though the molecular analysis agrees with the morphological analysis in general topology, they differ in the resolution within the slender-bodied subclade and the position of D. malabaricus .
Combined analysis of morphology and molecules
A combined analysis of 16 morphological characters and 246 variable sites in the DNA sequences from five genes yields 135 equally-parsimonious trees. The bootstrap consensus tree is shown in Figure 13 View Figure 13 (b). The combined analysis strongly supports the monophyly of Danio and its basal division into deep-bodied and slender-bodied subclades but it fails to resolve fully the relationships within the deep-bodied clade. The topology of the shallow-bodied clade is identical to that found in our molecular analysis ( Fig. 13a View Figure 13 ) and previous molecular studies ( Meyer et al., 1995; Zardoya et al., 1996), but differs from the topology derived only from morphological data. The topology of the deepbodied subclade derived from combined data is identical to that obtained when morphological data are analysed alone.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |