Gint gubanensis, Kovarik, Lowe, Just, Awale, Elmi & Stahlavsky, 2018
|
publication ID |
https://doi.org/ 10.1093/zoolinnean/zlac049 |
|
DOI |
https://doi.org/10.5281/zenodo.7194672 |
|
persistent identifier |
https://treatment.plazi.org/id/03AACF58-FFA7-1F22-FEF4-1709FECF94C9 |
|
treatment provided by |
Plazi |
|
scientific name |
Gint gubanensis |
| status |
|
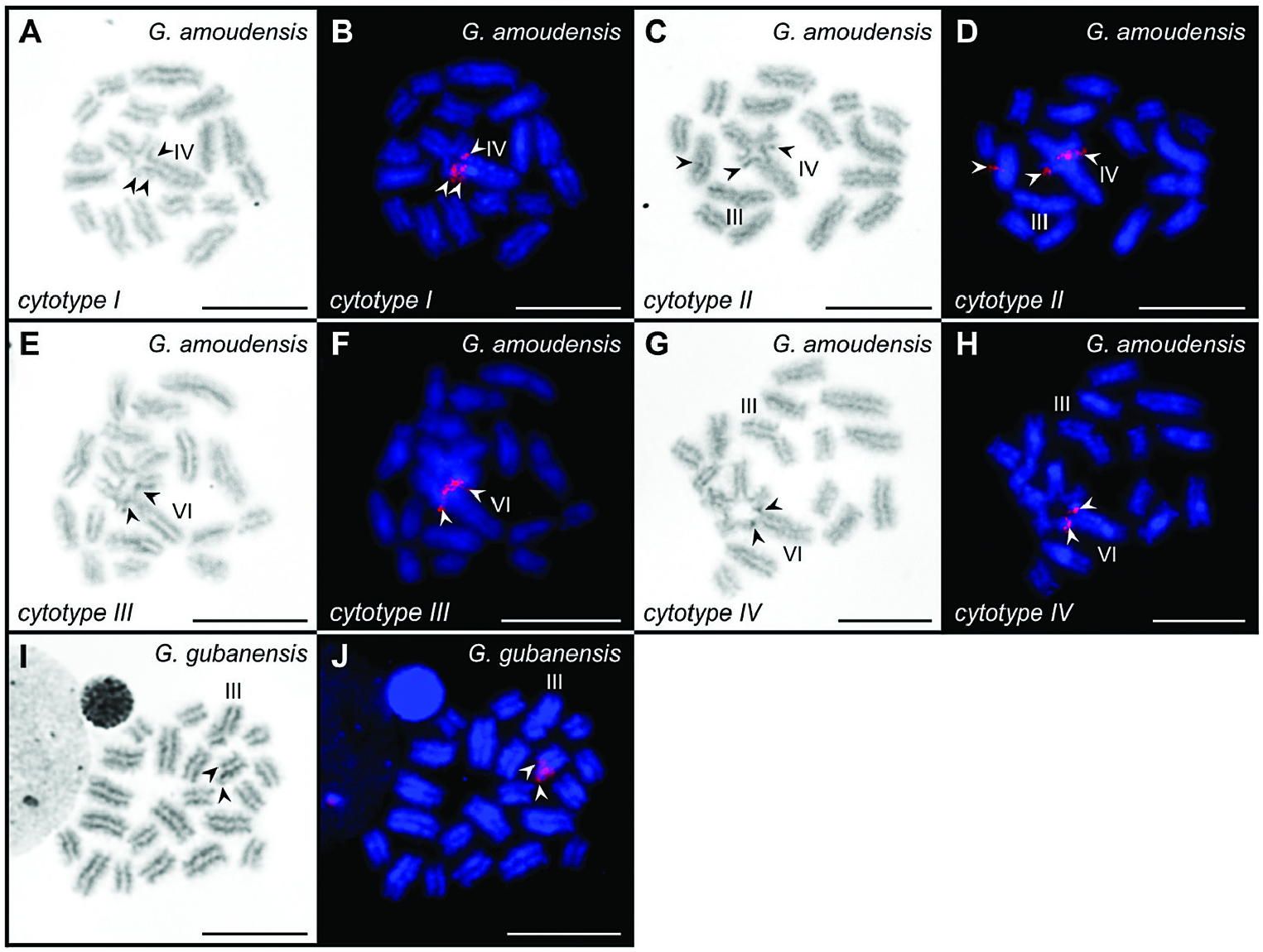
Karyotype of G. gubanensis comprises 2 n = 45 chromosomes, which decrease in length from 2.97% to 1.02% of DSL (Supporting Information, Fig. S2G View Figure 2 ; Table S2). Overall, 21 bivalents and a single trivalent, composed of chromosomes 7, 33 and 45, are detected in post-pachytene nuclei ( Fig. 3I View Figure 3 ; Supporting Information, Fig. S2G View Figure 2 ). One pair of 18S rDNA loci is found on a terminal part of the heteromorphic chromosome pair (chromosomes 26 and 44). A single large 18S rDNA cluster is located on the overhanging part of chromosome 26 (the larger chromosome of the heteromorphic pair) ( Fig. 3J View Figure 3 ; Supporting Information, Fig. S2G View Figure 2 ).
MTDNA DIVERSITY AND PHYLOGENY
The sequences for both 16S and COI were obtained from 22 Gint individuals. In G. calviceps , only the 16S gene fragment was successfully sequenced. The final alignment with a total length of 963 bp (16S: 358 bp; COI: 605 bp) consisted of 765 conserved sites, 194 variable sites and 172 parsimony-informative sites.
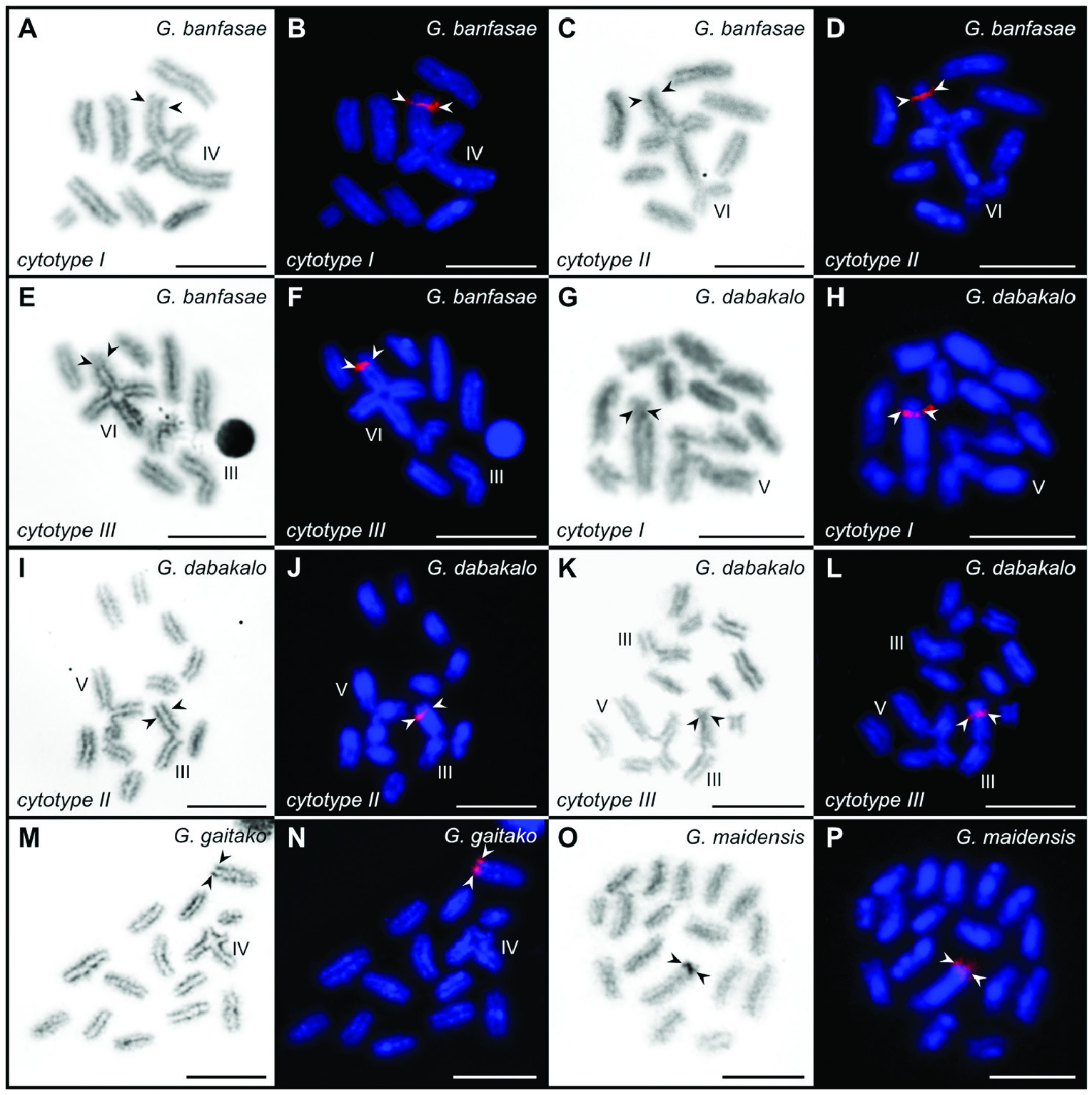
Based on COI data, Gint exhibits genetic variation at both inter- and intraspecific levels, with the pairwise genetic distances ranging from 3.28% to 11.52% between species, and from 0.00% to 2.64% within species (Supporting Information, Tables S3, S4). The haplotype network reconstruction showed a presence of 15 COI sequence haplotypes in Gint (Supporting Information, ( Fig. S3 View Figure 3 ; Table S3). Within the species studied, G. banfasae shows the highest within-location haplotype diversity (four haplotypes per locality), but with minor pairwise genetic differences (p -distance: ≤0.66%). In this species, all individuals of the same cytotype have distinct haplotypes. The haplotypes of cytotype I individuals (S1531 and S1534) are separated from one another by four mutational steps. Cytotype II (S1532) shares its haplotype with the cytotype III individual (S1530), which is one mutational step distant from the haplotype of the cytotype III specimen (S1533) (Supporting Information, Fig. S3 View Figure 3 ). Gint dabakalo , comprising three haplotypes, shows the highest intraspecific genetic variation with a p -distance of 0.17–2.64%. In this species, the genetic distance between individuals of cytotype I and cytotype II from the same locality is higher than between each of them and a specimen of cytotype III from a distinct locality (Supporting Information, Fig. S3 View Figure 3 ; Table S1). Gint maidensis , a species with a stable karyotype 2 n = 34, is represented by three haplotypes with a p -distance of 0.33–0.50%. Gintgaitako presentstwohaplotypes, where one haplogroup, formed by five specimens, is distant from the other by one mutational step (p -distance: 0.17). In G. amoudensis , individuals of cytotype II, cytotype III and cytotype IV share the same haplotype, whereas the specimen of cytotype I differs from this haplogroup by eight point mutations (p -distance: 1.49%).
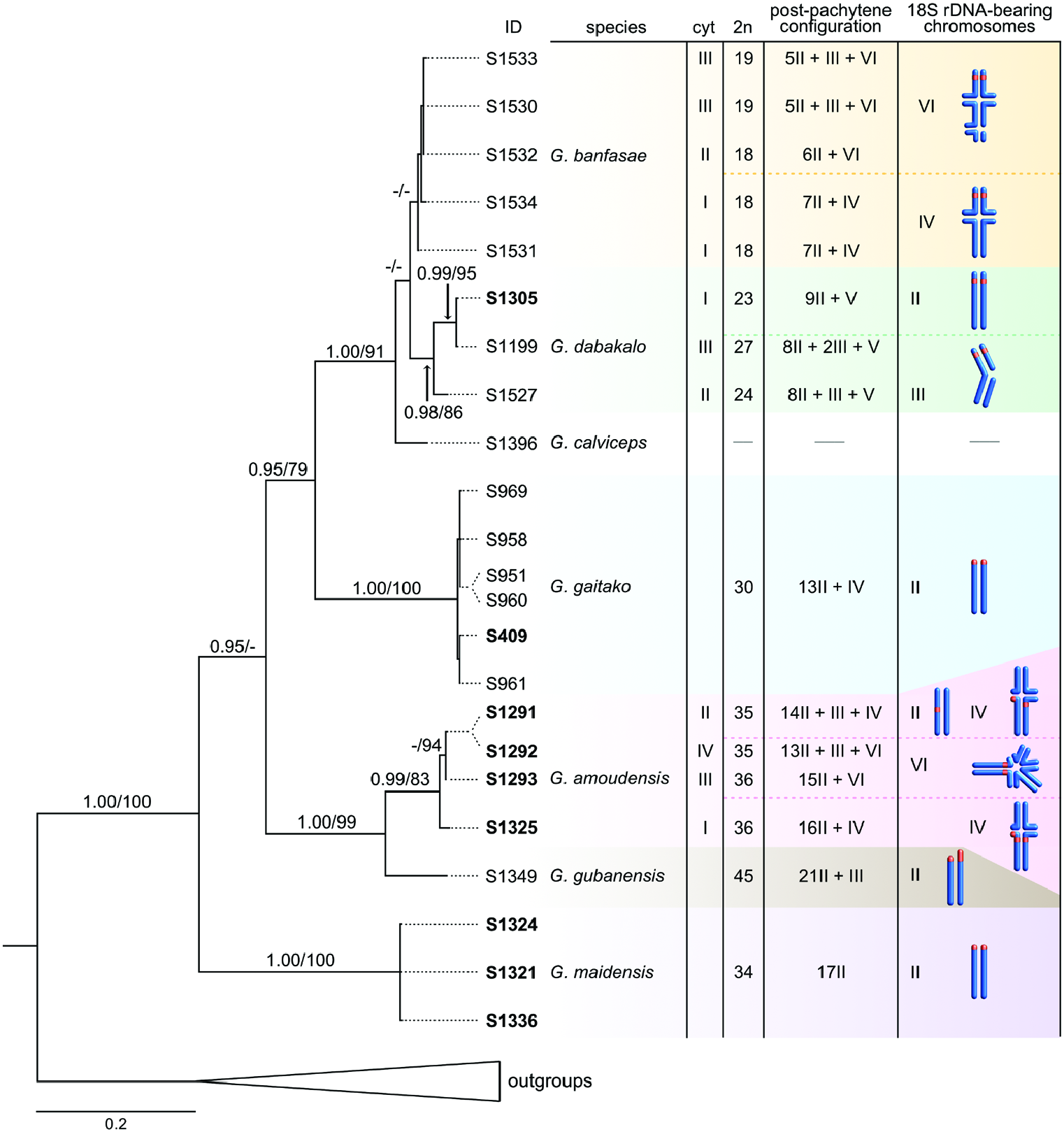
Both BI and ML mtDNA analyses provide similar tree topologies and nodal supports for clades. Phylogenetic reconstruction depicts Gint as a monophyletic group ( Fig. 5 View Figure 5 ). Most of the Gint species form well-supported clades [bootstrap support (bs) ≥ 83, posterior probability (PP) ≥ 0.98]. The monophyly of the G. banfasae specimens remains unresolved due to low support values. Gint maidensis forms an early-branching clade, which is sister to the clade comprising all of the remaining Gint species. This clade is further subdivided into two groups: (1) a clade in which G. gubanensis is a sister-species to G. amoudensis , and (2) a clade consisting of G. gaitako , G. calviceps , G. dabakalo and G. banfasae . In the latter, G. gaitako forms a sister-lineage to the clade comprising G. calviceps , G. dabakalo and G. banfasae . The mutual relationships between the species in this group remain unresolved due to low support values.
THE RECONSTRUCTION OF ANCESTRAL CHROMOSOME NUMBER AND POSITION OF 18S RDNA GENE CLUSTERS
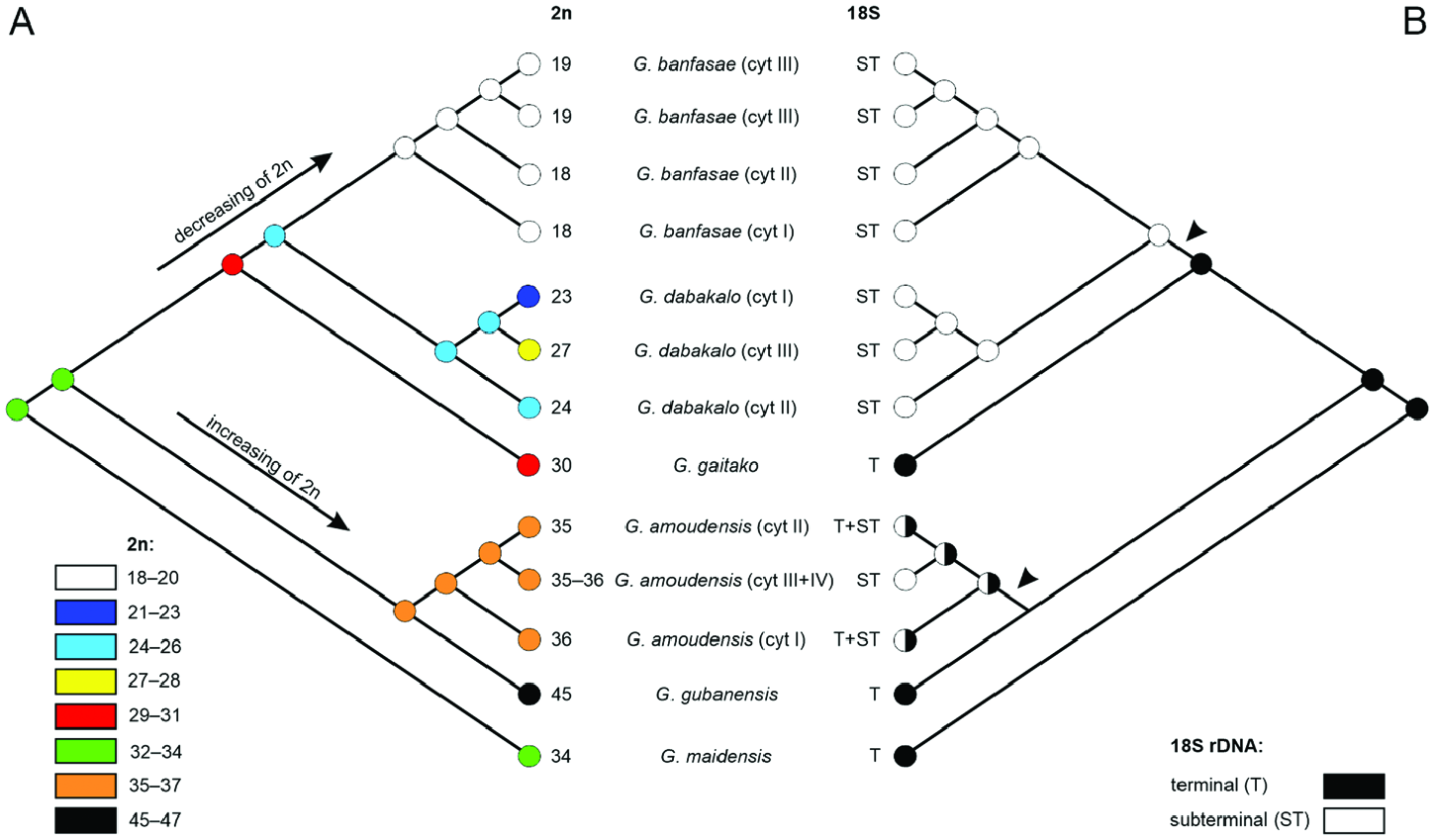
Maximum parsimony analysis inferred 2 n = 32–34 as the ancestral chromosome number for the genus Gint ( Fig. 6A View Figure 6 ). Two independent increases of 2 n were detected within Gint : (1) the clade containing all cytotypes of G. amoudensis and G. gubanensis , and (2) cytotype III within the G. dabakalo clade. When compared with the ancestral 2 n, a tendency of decreasing of 2 n from 30 to 18 chromosomes can be observed in G. gaitako , G. dabakalo and G. banfasae . Concerning the position of 18S rRNA gene clusters, maximum parsimony inferred their terminal position as the ancestral state for the genus Gint ( Fig. 6B View Figure 6 ). Two independent changes of the ancestral terminal position to a subterminal position were identified in G. amoudensis and in the clade containing G. dabakalo and G. banfasae .
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |