Rhododendron agastum, Balf. f. & W.W. Sm.
|
publication ID |
https://doi.org/10.1016/j.phytochem.2021.112655 |
|
DOI |
https://doi.org/10.5281/zenodo.8302427 |
|
persistent identifier |
https://treatment.plazi.org/id/0397C96B-FF9A-874F-473C-F9269309FCA3 |
|
treatment provided by |
Felipe |
|
scientific name |
Rhododendron agastum |
| status |
|
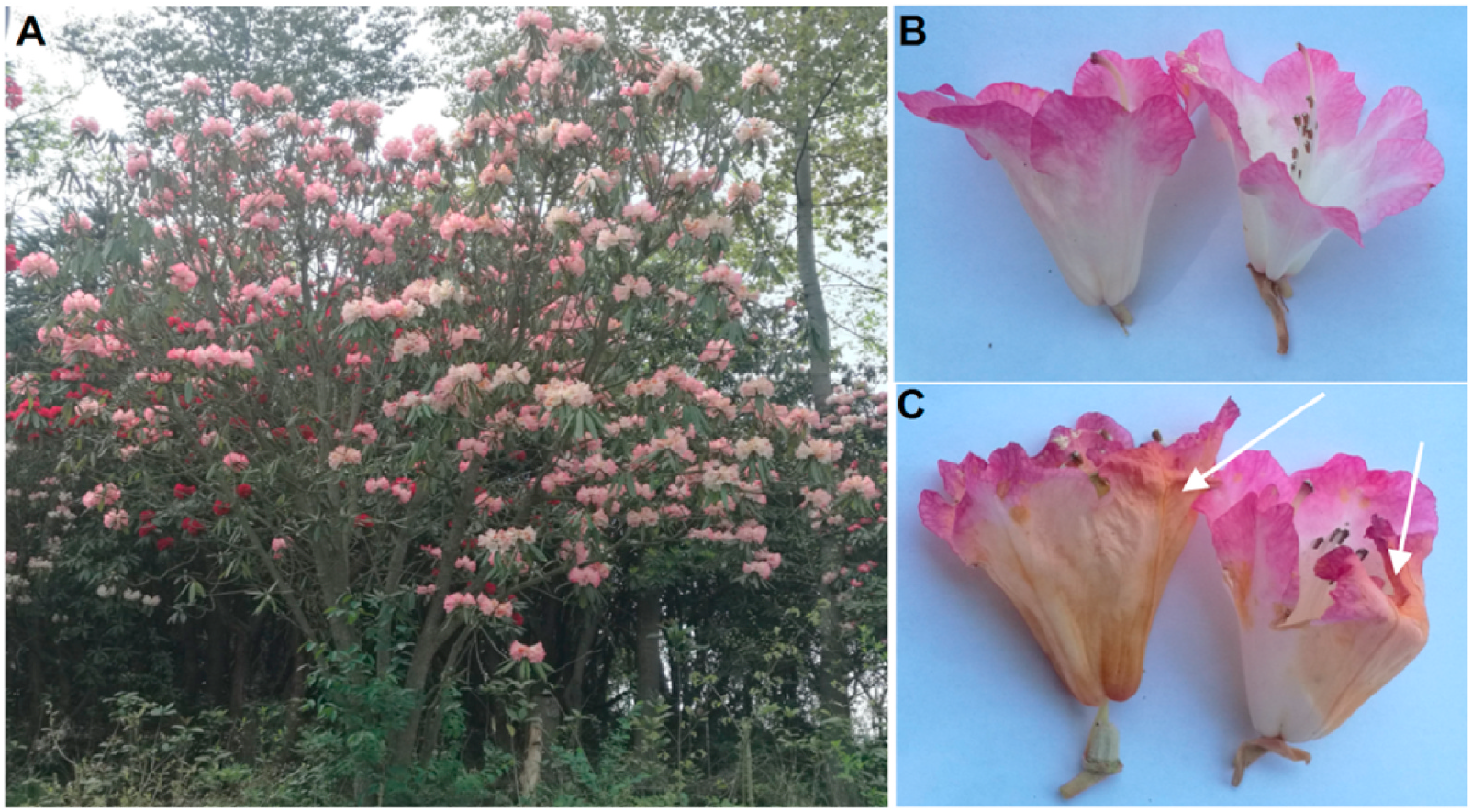
2.1. Metabolite identification in R. agastum View in CoL View at ENA flowers
The GC-TOF-MS and UHPLC-QTOF-MS/MS platforms, combined with annotation software and databases, were used to identify metabolites from healthy and petal blight flowers of R. agastum . GC-TOF-MS platform has advantages for the analysis of small volatile or semivolatile compounds, whereas UHPLC-QTOF-MS/MS platform is more suitable for phenolic compounds, flavonoids, and triterpenic acids ( Olmo-Garcia et al., 2018). To our knowledge, there have been no previous large-scale untargeted metabolomics studies of R. agastum that combined GC-TOF-MS and UHPLC-QTOF-MS/MS, especially for petal blight of Rhododendron species.
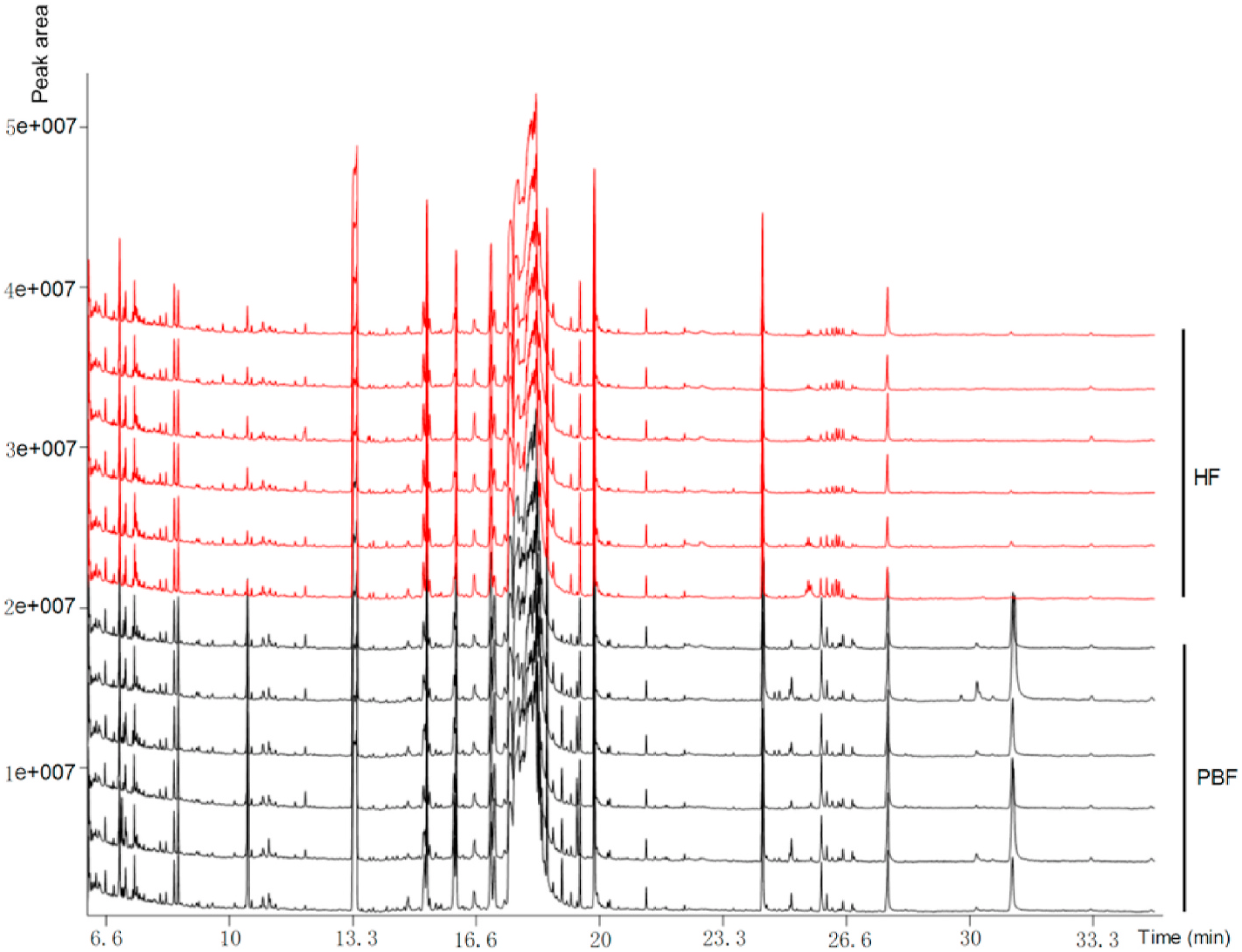
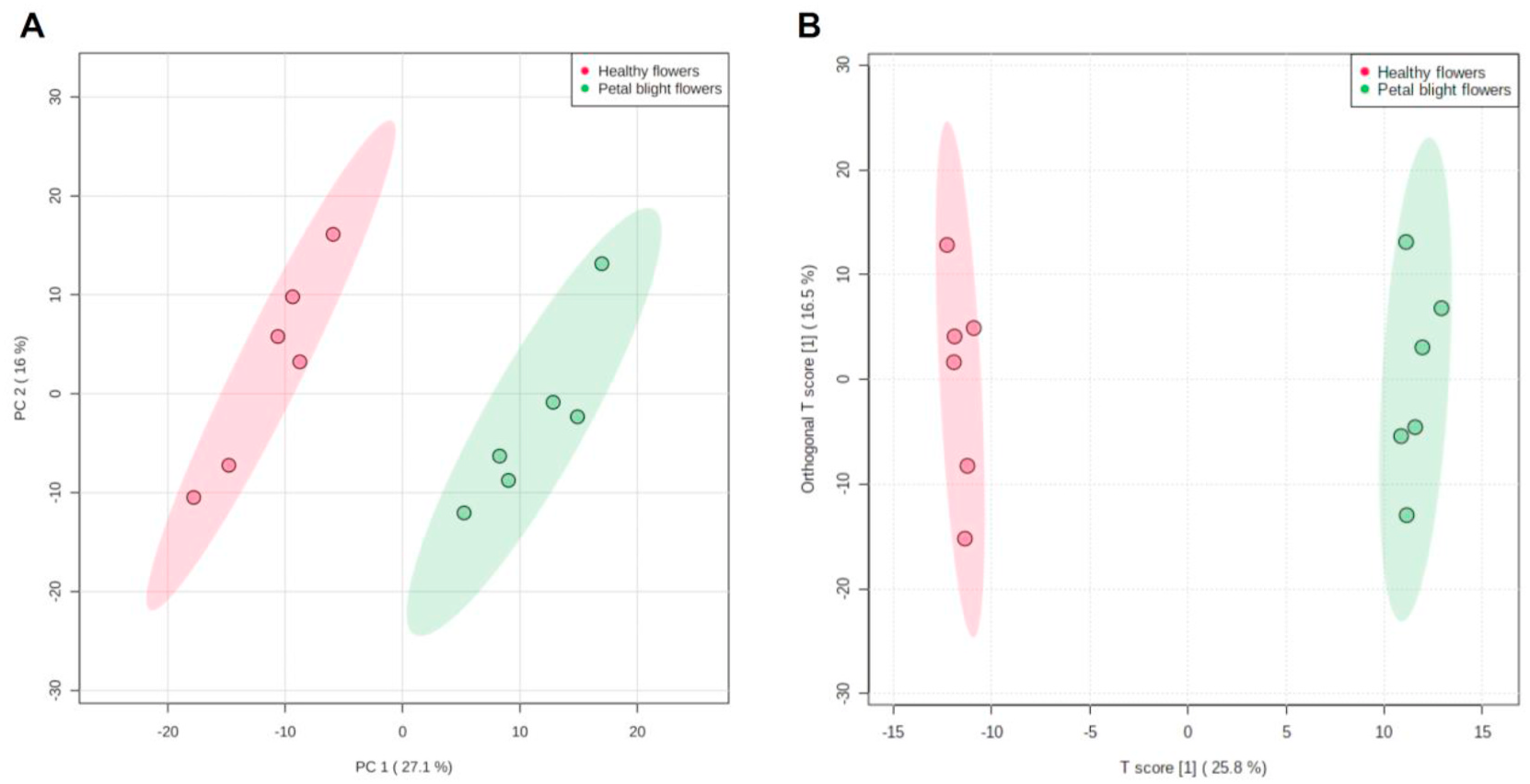
Differences in metabolites between healthy and petal blight flowers were evaluated by measuring six biological replicates. The GC-TOF-MS chromatograms of 12 samples from healthy and petal blight flowers showed good reproducibility, indicating that the run conditions were stable and reliable ( Fig. 2 View Fig ). The retention times and peak areas of six quality control samples also showed good repeatability during the experiment (Supplementary Fig. 1 View Fig ), indicating that the instrument itself was very stable. The relative deviation of the internal standard (saturated fatty acid methyl ester) added in the quality control sample was 8.21%, further verifying the system’ s stability. A total of 571 peaks were extracted, and 189 metabolites were tentatively identified based on mass spectrum match and retention index match.
Samples from healthy and petal blight flowers were also analyzed using the UHPLC-QTOF-MS/MS platform, and total ion chromatograms (TICs) were obtained in positive and negative ion mode (Supplementary Fig. 2 View Fig ). Four quality control samples also showed good repetitiveness during the experiment (Supplementary Fig. 3 View Fig ). The relative deviations of the internal standard (l-2-chlorophenylalanine) in the quality control samples were 6.66% and 2.37% in the positive and negative ion mode, respectively, indicating that the system was very stable. A total of 1731 and 1994 peaks were extracted, and 364 and 277 metabolites were tentatively identified in the positive and negative ion mode.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |