Panoquina albistriga O. Mielke, 1980
|
publication ID |
https://doi.org/10.11646/zootaxa.5271.1.3 |
|
publication LSID |
lsid:zoobank.org:pub:39D641B7-1800-4918-8E88-4EC5FF4BB56C |
|
DOI |
https://doi.org/10.5281/zenodo.7864355 |
|
persistent identifier |
https://treatment.plazi.org/id/F84A87F4-9B23-FFD6-FF3C-A768BE84FB93 |
|
treatment provided by |
Plazi (2023-04-24 10:09:35, last updated 2024-11-26 03:39:09) |
|
scientific name |
Panoquina albistriga O. Mielke, 1980 |
| status |
stat. nov. |
Panoquina eugeon (Godman & Salvin, 1896) View in CoL , reinstated status, Panoquina calna Evans, 1955 and Panoquina albistriga O. Mielke, 1980 , new status, and Panoquina eugeon minima de Jong, 1983 , new combination
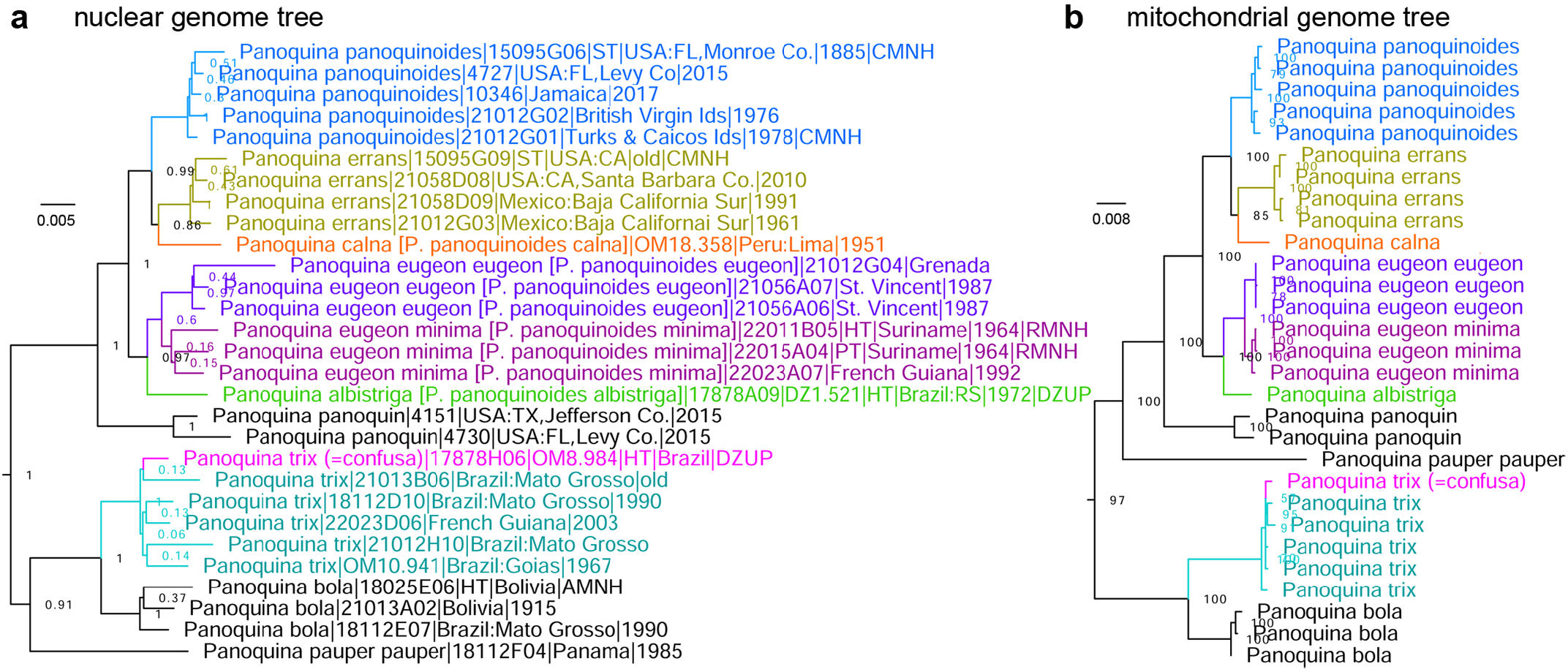
Genomic trees reveal that Panoquina panoquinoides (Skinner, 1891) (type locality in USA: Florida / Texas) is not monophyletic, and Panoquina errans (Skinner, 1892) (type locality in USA: California / Texas) originates within it ( Fig. 10 View FIGURE 10 ). We find that Panoquina panoquinoides calna Evans, 1955 (type locality in Peru) ( Fig. 10 View FIGURE 10 orange) is sister to P. errans ( Fig. 10 View FIGURE 10 olive, syntype sequenced) in both nuclear and mitochondrial genome trees, differing from it by 2.6% (17 bp) in COI barcodes and Panoquina panoquinoides albistriga O. Mielke, 1980 (type locality in Brazil: Rio Grande do Sul) ( Fig. 10 View FIGURE 10 green, holotype sequenced) is sister to Panoquina panoquinoides eugeon (Godman & Salvin, 1896) (type locality in Grenada) showing 0.9% (6 bp) in COI barcode distance (lower than expected from the nuclear genome divergence illustrated in Fig. 10 View FIGURE 10 ). Due to prominent genetic differentiation between these taxa, we propose to treat them as species: Panoquina eugeon (Godman & Salvin, 1896) , stat. rest., Panoquina calna Evans, 1955 , stat. nov. and Panoquina albistriga O. Mielke, 1980 , stat. nov. Sequencing Panoquina panoquinoides minima de Jong, 1983 (type locality in Surinam) ( Fig. 10 View FIGURE 10 maroon) demonstrates that it is sister to P. eugeon , stat. rest. ( Fig. 10 View FIGURE 10 purple) and clusters with it rather closely, especially in the mitogenome tree (COI barcode difference 0.76%, 5bp). Due to genetic and phenotypic similarities and proximity in locality we conservatively place it as a subspecies: P. eugeon minima de Jong, 1983 , comb. nov. pending more detailed research.
de Jong, R. (1983) Annotated list of the Hesperiidae (Lepidoptera) of Surinam, with descriptions of new taxa. Tijdschrift voor entomologie, 126, 233 - 268.
Evans, W. H. (1955) A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part IV. Hesperiinae and Megathyminae. The Trustees of the British Museum (Natural History), London, v + 499 pp., pls. 454 - 488.
FIGURE 10. Panoquina trees constructed from protein-coding regions in: a. nuclear genome, b. mitochondrial genome. Specimens are arranged in the same order in both trees and complete labels are given in panel a only. P. panoquinoides (blue), P. errans (olive), P. calna, stat. nov. (orange), P. eugeon eugeon, stat. rest. (purple), P. eugeon minima, comb. nov. (maroon), P. albistriga, stat. nov. (green), P. trix (cyan with its synonym P. confusa in magenta).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
SubTribe |
Calpodina |
|
Genus |