Reseda, L.
|
publication ID |
https://doi.org/10.1016/j.phytochem.2021.112658 |
|
DOI |
https://doi.org/10.5281/zenodo.8273672 |
|
persistent identifier |
https://treatment.plazi.org/id/66798798-FFA5-FFF9-634C-F89DFE66FECF |
|
treatment provided by |
Felipe (2023-08-17 21:56:31, last updated 2024-11-27 08:42:19) |
|
scientific name |
Reseda |
| status |
|
2.4. Glucosinolates in a distantly related genus, Reseda View in CoL View at ENA
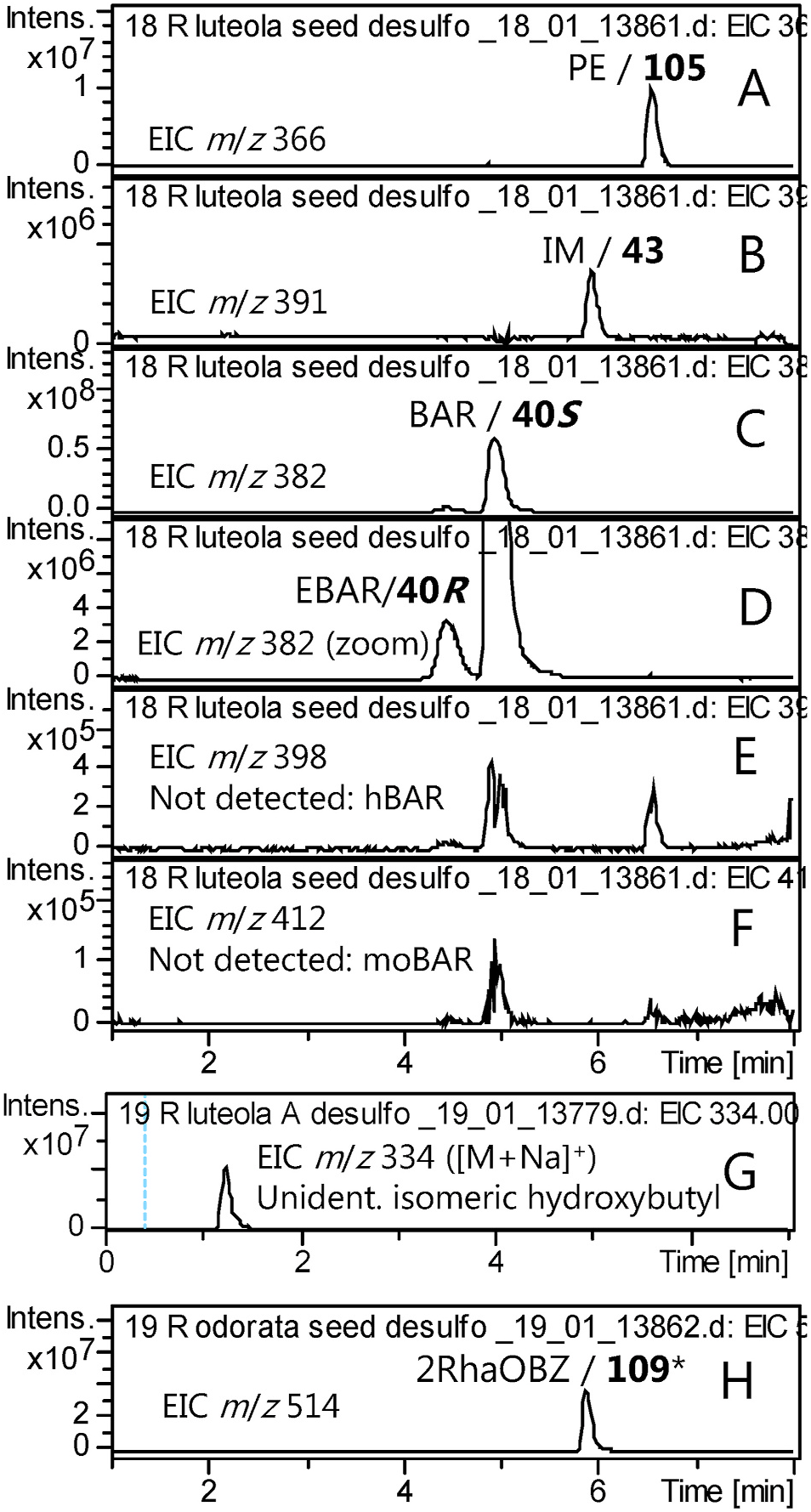
To provide a perspective of the diversity in the tribe Cardamineae , the species Reseda luteola (family: Resedaceae ) was investigated because the literature suggested the GSL profile to remarkably resemble that of B. vulgaris . The presence of BAR in R. luteola is well-established ( Kjaer and Gmelin, 1958; Bennett et al., 2004), despite the great evolutionary distance to Barbarea . We revisited this species in order to check for traces of the epimer EBAR and related GSLs. The major peaks were confirmed to be BAR, PE and IM, in seeds ( Fig. 9A–C View Fig ) and in leaves. A minor peak was conclusively identified as EBAR ( Fig. 9D View Fig ). However, hydroxy or methoxy substituted glucobarbarins were not detected ( Fig. 9E–F View Fig ). As these are available as references from Barbarea and Arabis spp. and known to be detectable and base line separated from the R. luteola peaks at our conditions ( Table 2 View Table 2 ; Olsen et al., 2016), their absence (above the limit of detection) could be concluded with certainty.
The level of EBAR in R. luteola seeds corresponded to ca. 5% of the sum of EBAR and BAR as estimated from peak intensities, but was relatively lower in leaves, ca. 1% of the sum of EBAR and BAR. The varying proportion of the epimeric glucobarbarins in seeds and leaves of R. luteola suggests that each epimer is due to a distinct genetic locus and biosynthetic enzyme, as is the case in B. vulgaris ( Liu et al., 2019a).
Putative constituents like BZ, 1mEt, 1mPr/2mPr and isomers of mBu and mPe were searched for in both leaves and seeds of R. luteola but not detected (results not shown). However, an apparent isomer of hydroxybutylGSL was detected ( Fig. 9G View Fig ), suggesting that use of amino acid precursors for seed GSL biosynthesis in R. luteola is not quite as restricted, to Trp and homoPhe, as in B. vulgaris , but include an aliphatic precursor.
We further investigated the apparent hydroxybutylGSL in R. luteola . Only trace-levels were found in seeds, but analysis of leaves revealed appreciable levels of the apparent hydroxybutylGSL ( Fig. 9G View Fig ), which eluted at 1.3 min and hence was not 1hmPr (30) but could be either Met-derived 4hBu ([26]) (not previously known from basal families) or Leuderived 2h2mPr (31), not available in our reference library at that time, or several yet unknown isomers. Ion trap MS2 was inconspicious and identical to both the MS2 of d [26] and d 30. We re-analyzed the sample using UHPLC-QToF MS/MS and found the high-resolution mass to be in agreement with a desulfo hydroxybutylGSL (found: 312.1114 (mean, N = 2), calc. for C 11 H 22 O 7 NS + ([M+H] +): 312.1112). This result was in agreement with a previous report of an unspecified hydroxybutyl GSL in leaves of R. luteola ( Griffiths et al., 2001) .
Neither BAR nor EBAR were detected in Reseda odorata seeds in the present study, in contrast to a previous report ( Bennett et al., 2004). The dominating dGSL peak at m / z = 514 was presumably due to the well-established 2RhaOBZ (109) ( Pagnotta et al., 2020). The dGSL d 109 showed a distinct MS2 compared to that from the isomer d 110 (Supplementary Fig. S1C View Fig ) and confirmed our ability to detect a wide range of structural types including glycosides ( Fig. 9H View Fig ). A minor peak of desulfo IM almost coeluted with desulfo 2RhaOBZ. A range of aliphatic GSLs including butyls and hydroxybutyls were searched for but not detected in seeds.
While both Reseda species accumulated IM, neither of the usual IM derivatives for tribe Cardamineae (Supplementary Fig. S1A View Fig ) were detected, although they were specifically searched for. No previous author has reported IM derivatives from the genus, even when roots were examined ( Pagnotta et al., 2020). However, roots should be investigated critically before concluding on presence or absence of substituted indole GSLs in this genus.
Bennett, R. N., Mellon, F. A., Kroon, P. A., 2004. Screening crucifer seeds as sources of specific intact glucosinolates using ion-pair high-performance liquid chromatography negative ion electrospray mass spectrometry. J. Agric. Food Chem. 52, 428 - 438. https: // doi. org / 10.1021 / jf 030530 p.
Griffiths, D. W., Deighton, N., Birch, A. N. E., Patrian, B., Baur, R., St ¨ adler, E., 2001. Identification of glucosinolates on the leaf surface of plants from the Cruciferae and other closely related plants. Phytochemistry 57, 693 - 700. https: // doi. org / 10.1016 / s 0031 - 9422 (01) 00138 - 8.
Kjaer, A., Gmelin, R., 1958. Isothiocyanates XXXIII. An isothiocyanate glucoside (glucobarbarin) of Reseda lueteola L. Acta Chem. Scand. 12, 1693 - 1694.
Liu, T. - J., Zhang, Y. - J., Agerbirk, N., Wang, H. - P., Wei, X. - C., Song, J. - P., He, H. - J., Zhao, X. - Z., Zhang, X. - H., Li, X. - X., 2019 a. A high-density genetic map and QTL mapping of leaf traits and glucosinolates in Barbarea vulgaris. BMC Genom. 20, 371. https: // doi. org / 10.1186 / s 12864 - 019 - 5769 - z.
Olsen, C. E., Huang, X. - C., Hansen, C. I. C., Cipollini, D., Orgaard, M., Matthes, A., Geu-Flores, F., Koch, M. A., Agerbirk, N., 2016. Glucosinolate diversity within a phylogenetic framework of the tribe Cardamineae (Brassicaceae) unraveled with HPLC-MS / MS and NMR-based analytical distinction of 70 desulfoglucosinolates. Phytochemistry 132, 33 - 56. https: // doi. org / 10.1016 / j. phytochem. 2016.09.013.
Pagnotta, E., Montaut, S., Matteo, R., Rollin, P., Nuzillard, J. M., Lazzeri, L., Bagatta, M., 2020. Glucosinolates in Reseda lutea L.: distribution in plant tissues during flowering time. Biochem. Systemat. Ecol. 90, 104043. https: // doi. org / 10.1016 / j. bse. 2020.104043.
Fig. 9. Extracted ion HPLC-MS chromatograms of desulfoglucosinolates prepared from glucosinolates (GSLs) in seeds (A–F) or leaves (G) of Reseda luteola and seeds of Reseda odorata (H). The three major peaks (A, B, C) represent PE, IM and BAR, much like many Barbarea spp. Focus on minor peaks (D) allowed conclusive identification of EBAR, confirmed by tR and the characteristic MS2 spectrum. A range of putative derivatives were not detected (E, F), but an unidentified hydroxybutylGSL was present (G), as was a known glycoside in R. odorata.
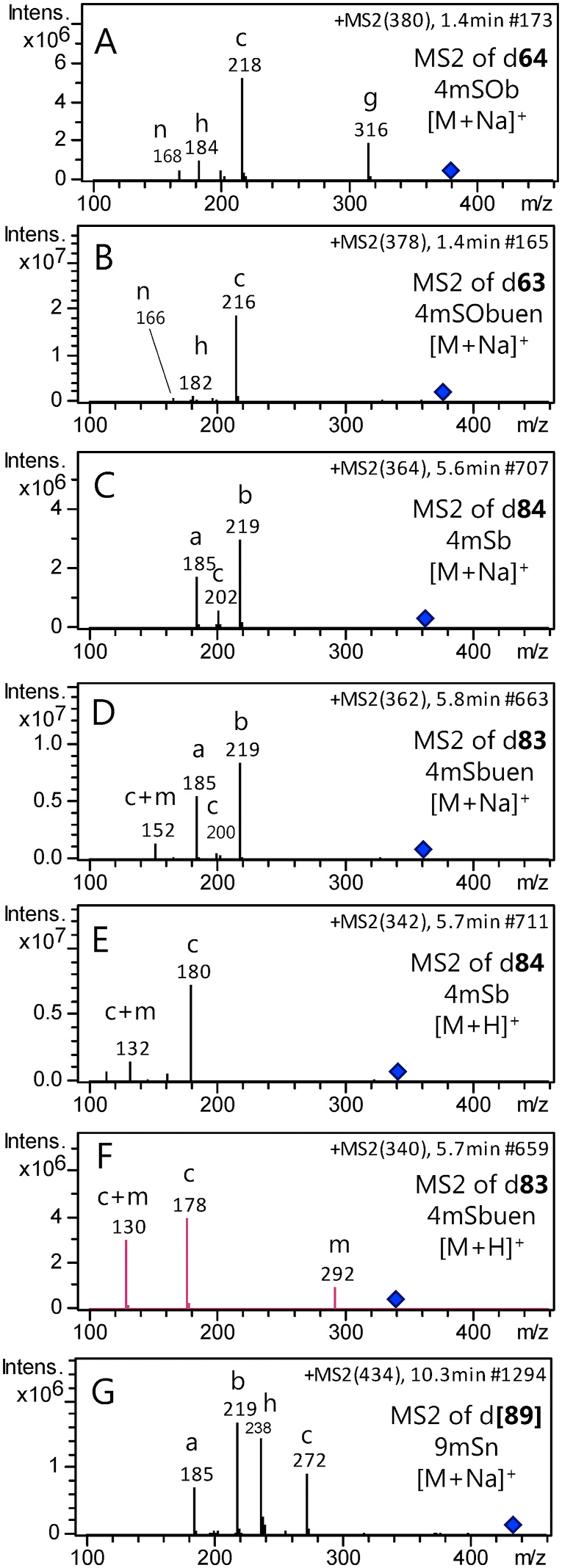
Fig. 1. MS2 spectra of pairs of desulfoglucosinolates with and without a side chain double bond. Four short chain desulfoglucosinolates were investigated, including Na+ adducts of all (A–D) and in addition H+ adducts of the methylthio substituted (E–F), as indicated in each spectrum. The desulfo derivative of the putative 9mSn ([89]), poorly characterized in the literature, was also investigated (G).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
1 (by felipe, 2023-08-17 21:56:31)
2 (by ExternalLinkService, 2023-08-17 23:07:39)
3 (by ExternalLinkService, 2023-08-22 13:13:49)
4 (by ExternalLinkService, 2023-08-22 14:16:28)
5 (by ExternalLinkService, 2023-08-22 15:25:25)
6 (by ExternalLinkService, 2023-08-28 15:45:20)
7 (by plazi, 2023-11-10 03:57:00)