Phylloscopus emilsalimi, Rheindt & Prawiradilaga & Ashar & Lee & Wu & Ng, 2020
|
publication ID |
https://doi.org/ 10.5281/zenodo.3608758 |
|
publication LSID |
lsid:zoobank.org:pub:8114B399-C68D-43C2-B6D3-B51AA898431E |
|
DOI |
https://doi.org/10.5281/zenodo.3610545 |
|
persistent identifier |
https://treatment.plazi.org/id/03F587A7-FFBF-8F31-FF12-7026FE3BF869 |
|
treatment provided by |
Plazi |
|
scientific name |
Phylloscopus emilsalimi |
| status |
sp. nov. |
SM6:
Phylloscopus emilsalimi , species nova
(Taliabu Leaf-Warbler;
urn:lsid:zoobank.org:act: E77116 View Materials DC-D83F-4C3C-A472-519B99D2843F
) Frank E. Rheindt, Dewi M. Prawiradilaga, Hidayat Ashari, Suparno, Nathaniel S. R. Ng
Holotype GoogleMaps
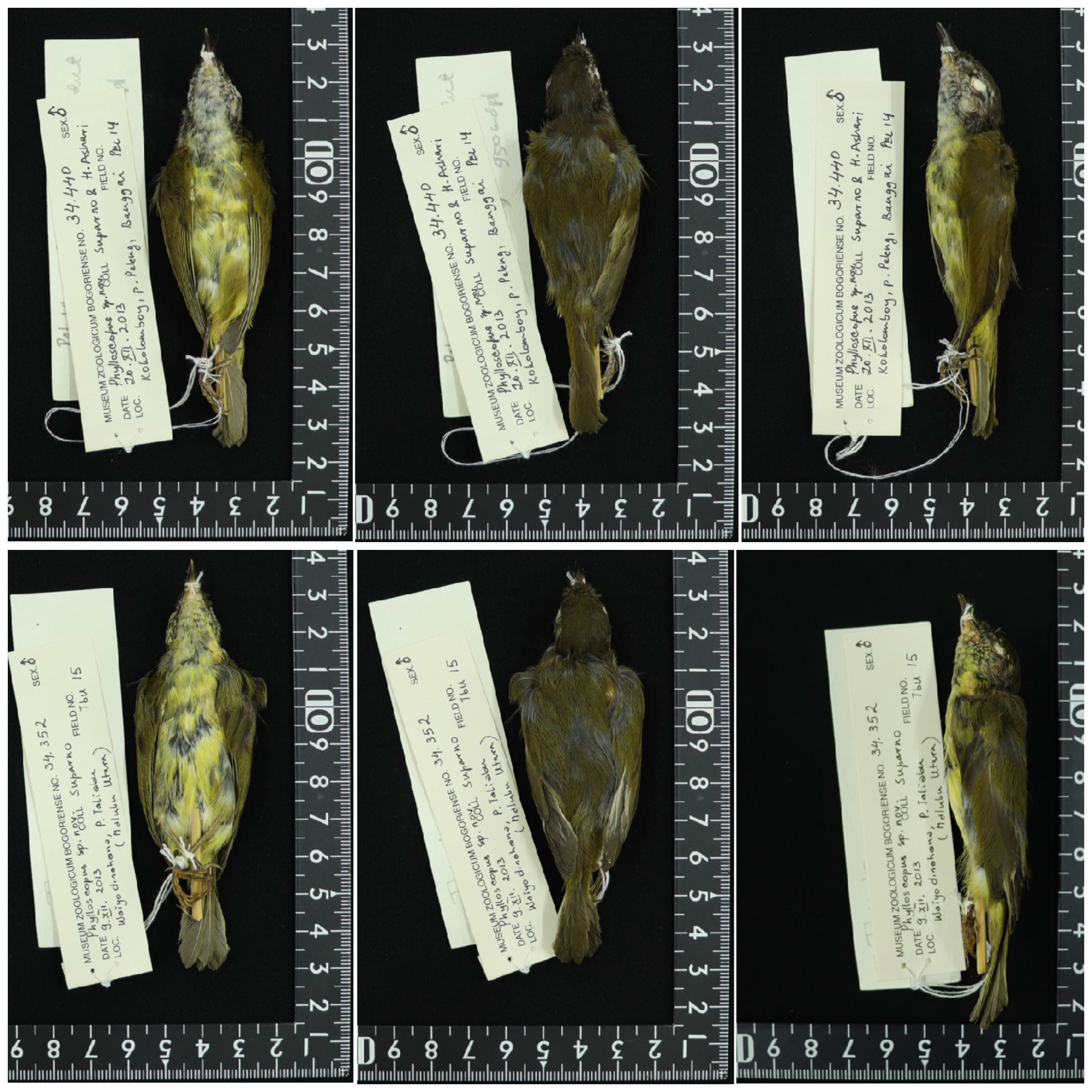
MZB.Ornit.34.352 ( fig. S8 View Fig ); adult male collected 9 Dec 2013 at Waiyo dinahana Camp GoogleMaps (~ 1200m) above the village of Wahe on Taliabu Island GoogleMaps (S 01⁰ 47.614 '; E 124⁰ 48.216 '). Collected by the Rheindt / LIPI field party, including tissue samples from breast muscle and liver; skin prepared by Suparno; field number Tbu15; no molt; medium to high fat; weight 10g; wing length 6.8cm; wing spread 17.6cm; total length 10.2cm; bill 1.35cm; tail 4.1cm; tarsus 2.5cm.
Description of holotype
The crown (from forehead to nape) is dark brownish-olive (5Y 4/6), delimited below by a narrow long lemon supercilium (5Y 8/4) from bill base to nape side. Partial eye ring formed by small white feathers around the eye is broken by a dark brownish-olive eyestripe (5Y 3/3). Black rictal bristles. Lemon ear coverts down to malar and moustachial regions (5Y 9/10) grizzled with dusky-olive feather edging, making for a mottled facial appearance. Upperparts from mantle and scapulars to rump are mid-olive (7.5Y 4/6). Remiges are distinctly darker- olive (5Y 3/2) but have paler olive outer edging concolorous with mantle. Upperwing coverts have outer webs largely concolorous with mantle and inner webs concolorous with remiges. Rectrices are dark olive above (5Y 3/2) with brighter basal edging roughly concolorous with mantle, and have a more silvery sheen below (5Y 5/1). Underparts from chin to vent are largely lemon to sulphur yellow, with white feather bases creating a more lemon-yellow hue on throat, breast and belly (7.5Y 8/8) and olive feather tips (5Y 4/4) generating a more olive- mottled appearance on flanks, while the undertail coverts are largely uniform lemon (5Y 9/12).
Iris brown; bill dark horn with base of lower mandible much paler pink-horn. Tarsus and toes on live bird reported as pink-horn with paler yellowish toepads.
Diagnosis
Akin to the newly described species from Peleng, P. emilsalimi from Taliabu is a small Phylloscopus typical of the P. poliocephalus leaf-warbler radiation from across Australo- Papua and Wallacea. It is most notable for its entirely lemon underparts (including throat), and the lack of a central crown stripe and wingbars. It differs from other members of the complex in bioacoustic traits in addition to a number of important plumage features:
The new species P. emilsalimi from Taliabu differs strikingly from the newly described species from Peleng ( P. suaramerdu , see SM5), which is the geographically most proximate member of the radiation, in its much yellower (versus white) supercilium, facial grizzling, and throat. P. emilsalimi also has less of a mottled appearance on the breast, a greener crown, and paler bill and tarsus.
The new Taliabu species is most similar to P. nesophilus from large parts of montane Sulawesi, but differs in its paler bill and tarsus, its duller olive upperparts, and its less olive- mottled appearance on the breast. Specifically, it can be told from the geographically most proximate populations of P. nesophilus (from eastern, southeastern and central Sulawesi) by its paler yellow underparts coloration caused by an arrangement in which most feathers are predominantly white with strong yellow edging (versus all-yellow feathers, often with olive edges, in eastern Sulawesi birds). The new species is very distinct from P. sarasinorum in lacking both a pale central crown stripe and extensive white inner webs of outer rectrices.
P. emilsalimi generally differs from remaining members of the P. poliocephalus complex by a combination of features. In particular, it differs from P. poliocephalus giulianettii (Central Papua) in its lack of wingbars and lack of central crown stripe; from P. p. poliocephalus (West Papua) in its lack of wingbars, a yellower (versus more whitish) throat, a more olive (versus greyish) crown, and a much shorter, yellower (versus long and white) supercilium; from P. ceramensis (Seram) in its lack of well-defined wingbars, lack of central crown stripe, a yellower (versus white) supercilium, and a more olive tinge to the crown and auriculars (versus a darker grayish-brown head coloration); from P. everetti (Buru) in its lack of wingbars, slightly yellower throat, yellower (versus white) supercilium and olive (versus grayish) crown; from P. waterstradti (North Moluccas) in its yellower (versus white) throat, lack of wingbars (present in Obi population), an olive (versus grayish) head coloration and yellower supercilium; from P. floris (Flores) in its non-contrasting olive (versus contrastingly grey) crown, a yellower (versus white) supercilium, and less intensely yellow underparts; and from P. presbytes (Timor) in its lack of a central crown stripe, a yellower and shorter (versus long white) supercilium, and a yellower and less contrasting throat (versus a contrasting white throat).
Etymology
We are pleased to name this new species after Prof Emil Salim, former Minister of Environment of the Republic of Indonesia and eminent environmentalist whose actions have contributed to more stringent international guidelines regulating extractive industries and mining, thereby benefitting many countries’ natural communities, including Indonesia.
Individual, sex and age-related variation within the taxon
A second specimen (MBZ.Ornit.34.387, an adult female) is very similar to the holotype, perhaps with a slightly darker crown, suggesting that this species may be sexually monomorphic.
History of discovery
A University of East Anglia expedition by Davidson et al. ( 64) was the first to find a resident population of Phylloscopus leaf-warblers on Taliabu, which they encountered commonly in montane forest and which they considered to represent an undescribed taxon. Rheindt ( 48) then studied this leaf warbler during his visit between 4-18 April 2009, and we found them again during our expedition between 6-16 Dec 2 013 and collected two ( 19).
Distribution and status
The new warbler inhabits disturbed and undisturbed montane forest on Taliabu from 700m up to probably the highest elevations above 1,400m ( 19, 48, 64). It may also occur on the large neighboring island of Mangole, which rises to 1,127m and should harbor enough area of montane forest habitat above 700m to sustain a population. However, a lack of fieldwork in the mountains of Mangole means that there is no information available on its montane avifauna. The smallest main constituent of the Sula Archipelago, the densely populated island of Sanana, is unlikely to sustain any Phylloscopus leaf-warblers as it barely reaches above 600m, which seems to be too low for this species. The species is replaced on the Banggai Archipelago by the newly described P. suaramerdu (see SM5), while P. sarasinorum and P. nesophilus replace it in various parts of Sulawesi. Given its more relaxed elevational requirements and its tolerance for disturbed habitat, this species is probably not as immediately endangered as some of the other montane Taliabu avifauna.
Taxonomic rationale (combined for P. suaramerdu and P. emilsalimi )
The leaf-warbler genus Phylloscopus constitutes a large Old World radiation of small passerines that is exceptionally well-studied in phylogenetic terms (10 6-1 09). Recent demonstrations of genus paraphyly in combination with Seicercus [summarized in (10 8)] have prompted a variety of new generic treatments, including a division into two re-defined genera [ Seicercus and Phylloscopus ; see ( 57)], but we here follow the latest phylogenetic treatise that suggests a merger of all Seicercus and Phylloscopus into a single genus Phylloscopus (10 8).
Despite the genus’s well-known evolutionary history, a sizeable sub-clade of Phylloscopus from the Australasian (including Indonesian) Archipelago has largely been omitted from modern molecular insights. Notable exceptions include the first-ever example of a new avian species description ( P. rotiensis ) partly based on genome-wide DNA data from the Timor region (11 0). However, most island populations of Phylloscopus in this region have been merged into a limited number of ‘wastebasket species’, e.g. Mountain Leaf Warbler P. trivirgatus and Island Leaf Warbler P. poliocephalus , although vocal and plumage differences among them often far exceed those documented among the better- studied continental groups of Phylloscopus (8 0). As a consequence, some modern treatments [e.g. ( 57)] have adopted a ‘yardstick approach’ ( 70) in which pairwise plumage and vocal differences between archipelagic taxa that are comparable in magnitude to those between better-studied continental species pairs (or exceed them) are interpreted as being indicative of species status. In this way, a number of highly distinct island taxa have been elevated to species level, such as Buru Leaf Warbler P. everetti, Seram Leaf Warbler P. ceramensis [a treatment that had previously been suggested on the basis of vocal and plumage comparisons; see ( 80)], and a number of additional Wallacean taxa. Our description of P. suaramerdu and P. emilsalimi follows this framework of affording species rank to taxa when vocal and plumage characters indicate deeper divisions. We also bring genome-wide DNA data to bear on the taxonomic classification of these two new species. However, following the yardstick approach as applied under the Biological Species Concept, we believe that biological characters, especially bioacoustic ones, will be the most important indicators when conferring species status in Phylloscopus leaf-warblers.
Plumage evidence: As for plumage characters, P. suaramerdu and P. emilsalimi differ from each other and from surrounding taxa in a number of important traits (see Diagnosis for each taxon) which are far more salient than the plumage differences of often identical-looking species pairs on the Asian continent that are otherwise vocally and phylogenetically deeply diverged [e.g. the three members of the Arctic Warbler P. borealis trio; (11 1)]. Most importantly, P. suaramerdu has a contrastingly white throat and supercilium and a grey-and-white grizzled auricular patch where all other surrounding taxa are much more yellowish, rendering it instantly recognizable in large series of specimens. In contrast, P. emilsalimi is superficially reminiscent of Sulawesi Leaf Warbler P. nesophilus in its overall more yellowish coloration, displaying a phenotypic leapfrog pattern that has been documented from the Taliabu-Peleng-Sulawesi island trio on multiple previous occasions (50, 7 2). However, this superficial resemblance notwithstanding, it has important distinctions in bare-parts coloration and overall pigmentation (see Diagnosis), apart from obvious vocal differences.
Bioacoustic evidence: In bioacoustic terms, the song of P. suaramerdu is characterized by melodious jumbles of 1–1.5sec, mostly at 2000–6000 Hz, with short individual high notes between 5000–7000 Hz occasionally interspersed. Song motifs generally lack repetitive element sequences or trills ( fig. S9 View Fig ). In contrast, the song of P. emilsalimi is characterized by 1.5–3sec strophes mostly between 2000–6500 Hz (typically of a wider frequency range than P. suaramerdu ) that constitute a melodious jumble often followed by a final repetitive phrase, which can sometimes be omitted ( fig. S9 View Fig ). In this respect, P. emilsalimi is slightly more reminiscent of the East Sulawesi population of P. nesophilus than P. suaramerdu from Peleng (despite the geographic proximity of the latter two). The P. nesophilus population on Gunung Tumpu (East Sulawesi) is often characterized by strong repetitive series at a very broad frequency range (2500–7000 Hz), mostly preceded by a variable but simple melodious series of elements between 2000–5000 Hz and a brief nasal note separating this variable series from the repetitive series ( fig. S9 View Fig ). Depending on the presence or absence of components, the song rendition (=motif) can be 1.5–4sec long. In summary, the songs of these three forms ( P. suaramerdu , P. emilsalimi and P. nesophilus ), while highly variable, are predictably different and strongly indicative of species status when put into the context of song differences among other Phylloscopus species (10 7).
Genomic evidence: We generated a set of ~4,100 SNPs and a genome-wide sequence alignment spanning almost 3 million base pairs (bp), along with an alignment of mitochondrial cytochrome- b (cyt- b) sequences, to shed light on the differentiation of Phylloscopus leaf-warblers from Sulawesi, Peleng and Taliabu. Details on laboratory and analytical methods are given in separate Methods paragraphs below.
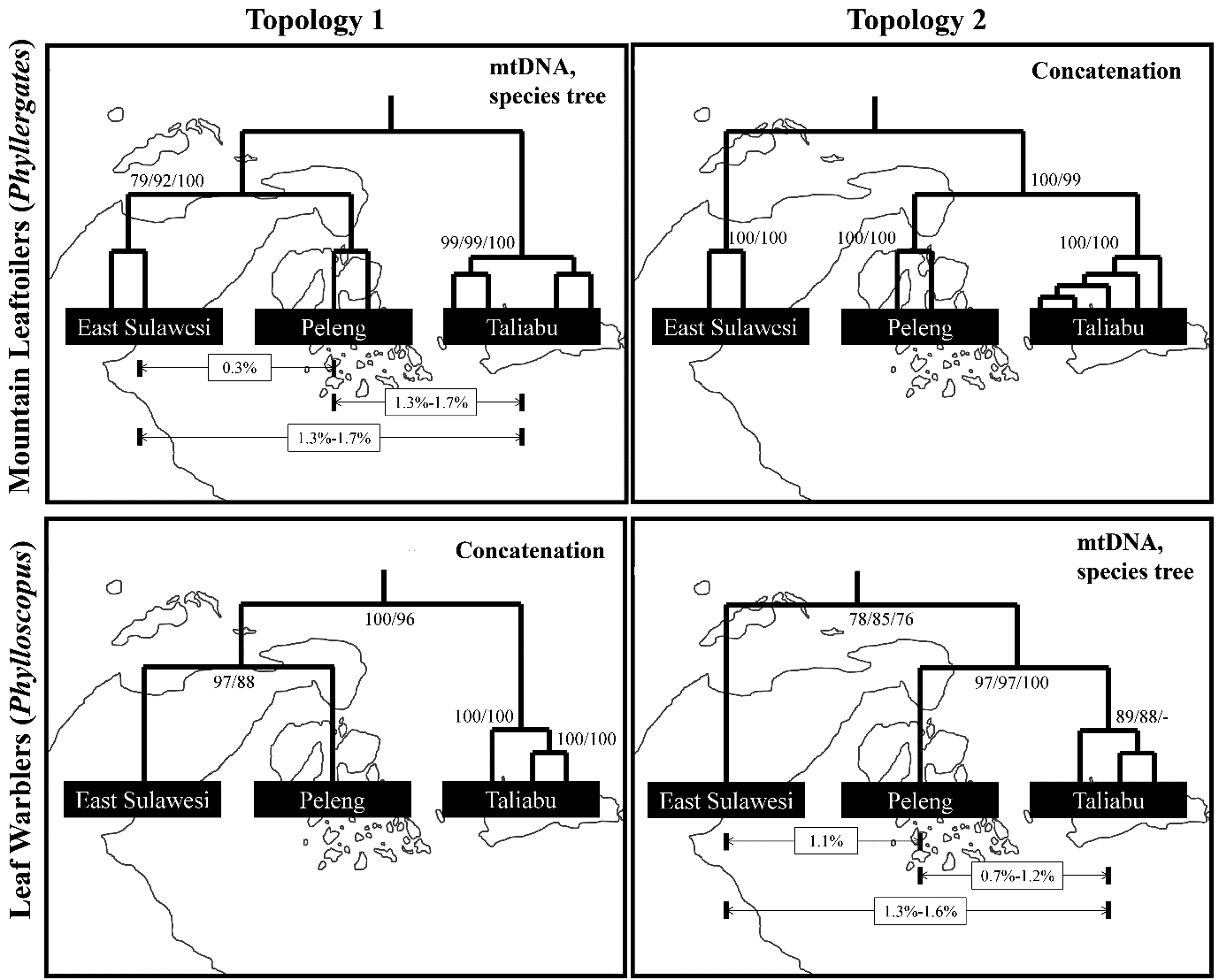
Each of the three taxa ( P. nesophilus from East Sulawesi, P. suaramerdu from Peleng and P. emilsalimi from Taliabu) formed a spatially separate cluster in population-genomic space based on ~4100 genome-wide SNPs ( fig. S10 View Fig ). The main division according to genomic PCA occurred between P. emilsalimi versus the other two taxa (PC1 in fig. S10 View Fig ). The same relationship was strongly reflected in maximum-likelihood (ML) analysis of concatenated genome-wide sequence alignments of almost 3 million base pairs length as well as in maximum-parsimony (MP) analysis of a concatenated set of ~4100 genome-wide SNPs (topology 1 in fig. S11 View Fig ). In contrast, mitochondrial cyt- b gene analysis and species-tree inference of genome-wide SNPs recovered a topology in which P. emilsalimi and P. suaramerdu were strongly supported sister lineages, with P. nesophilus being more basal (topology 2 in fig. S11 View Fig ). Depths of uncorrected pairwise sequence divergence (as determined by cyt- b distances) were comparatively low across all three leaf-warblers (0.7-1.6%; fig. S11 View Fig ).
In summary, our phylogenomic analyses demonstrated considerable incongruence among tree topologies, including a strong mito-nuclear discordance that calls into doubt the reliability of mitochondrial divergence values in this dataset. For instance, the deepest genomic divergence was detected between P. emilsalimi and the other two species (PC1 in fig. S10 View Fig ), whereas on mitochondrial cyt- b evidence P. emilsalimi displayed some of the most shallow divergences from the other taxa ( fig. S11 View Fig ). The use of mitochondrial divergences in species delimitation suffers from multiple confounding biases (79, 10 0, 10 1), especially when dealing with complexes containing closely-allied allopatric taxa that have a propensity for secondary gene flow (11 2-11 4). The best explanatory hypothesis for the pronounced mito- nuclear discordance in this dataset is that occasional overwater dispersal followed by hybridization and gene flow may have led to the introgression of foreign alleles in one or the other species, obfuscating clean branching patterns. Gene flow during sporadic hybridization is known to artifactually reduce mitochondrial divergences, masking deeper levels of genomic differentiation ( 79). Hence, introgression can account for the presence of relatively low mitochondrial divergences in a group of species characterized by comparatively deep genomic divergences and pronounced levels of bioacoustic differentiation [( 48, 49), see data above and fig. S9 View Fig ].
Methodology
Laboratory procedures: We extracted DNA from the fresh muscle tissue of the holotype of P. suaramerdu and the three specimens collected by us (including the holotype) of P. emilsalimi , along with one individual of P. nesophilus that we had collected on Mt Tumpu on the eastern peninsula of Sulawesi ( 19). Additional tissue samples were obtained from the American Museum of Natural History, New York (AMNH): three samples of P. sarasinorum from South Sulawesi and an outgroup sample of P. poliocephalus pallescens from the Solomon Islands. The AMNH samples were only used for phylogenomic rooting. DNA extraction was performed using the Exgene Clinic SV kit (GeneAll Biotechnology, Seoul) following the manufacturer’s instructions. We eluted the DNA in two consecutive rounds of 100μl molecular grade water. We prepared double-digest restriction enzyme associated DNA sequencing (ddRADSeq) libraries using the same protocol as Garg et al. (11 5) with EcoRI and MspI as restriction enzymes. We quantified DNA and library product concentrations using the Qubit 2.0 Broad Range DNA Assay (Invitrogen, California), and used a Fragment Analyzer (Advanced Analytical Technologies, Iowa) to perform final quality checks on the library product. The genomic libraries were sequenced on an Illumina HiSeq 3000 platform at the Singapore Center for Environmental Life Science Engineering. Filtering and demultiplexing of the genomic data in STACKS 1.34 followed the same general procedures as detailed by Ng et al. (7 2), and are the result of comprehensive trials using a wide variety of different parameters and settings at each step of the analysis. DNA data is deposited with Genbank at BioProject accession number PRJNA566263.
SNP calling and genomic supermatrix generation: Reference genome alignment was performed using BWA 0.7.12 (9 1) and samtools 0.1.19 (http://samtools.sourceforge.net) (9 2, 9 3) on the Greenish Warbler ( Phylloscopus trochiloides viridanus ) genome (11 6), the most closely related available full genome. We performed SNP calling in STACKS 1.34 as per Ng et al. (7 2). We required loci to be present across all individuals, and retained one random SNP per locus to exclude obviously linked SNP loci. Subsequently, we used PLINK 1.9 (11 7) and Bayescan 2.1 (11 8) to filter SNPs for linkage disequilibrium and signs of selection. Data format manipulation and conversions were performed with PGDSpider (1 19). The final filtered dataset totaled 4,104 SNPs. In addition, we created concatenated SNP datasets for each group for use in downstream maximum parsimony analysis.
Following the procedures used by Ng et al. (7 2), we generated a concatenated read supermatrix without missing data using pyRAD (12 0). We stipulated a minimum required locus coverage of 20, and kept the clustering threshold within and between reads at the default of 0.88. The final concatenated read supermatrix amounted to 2,861,661 bp in length, generated from 19,985 loci.
Mitochondrial sequence generation: We sequenced cyt- b using the primers H16065 (12 1) and L14995 (12 2), with 40 cycles of PCR performed at an annealing temperature of 72°C. We conducted PCR product cleanup with ExoSAP-IT ® and used the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc., Foster City, CA) for cycle- sequencing. Sequencing was carried out on an Applied Biosystems 3130xl Genetic Analyzer using capillary electrophoresis. We assembled cyt- b sequences with CodonCode Aligner 4.1 and aligned them with ClustalW (12 3). The final cyt- b alignment was 1,100bp long, indel- free and fully translatable. Mitochondrial sequences have been deposited with Genbank at accession numbers MN518858 View Materials - MN518862 View Materials .
Population structure inference: In order to test for the presence of geographic structuring across East Sulawesi, Peleng, and Taliabu, we applied principal component analysis (PCA) to the genomic SNP data. PCA was performed using ade4 (12 4) and factoextra (12 5), both available as R packages (12 6).
Mitochondrial tree inference: We constructed phylogenetic cyt- b trees using two methods: maximum parsimony (MP) and maximum likelihood (ML). We ran MP analyses using the seqboot, dnapars, and consense algorithms in PHYLIP v3.695 (12 7), performing 10,000 bootstrap replicates. For ML analyses, we first used jModelTest v2.1.6 ( 97) and determined that the Jukes-Cantor model (12 8) was the best performing substitution model. We then ran RAxML v8.2.9 ( 96) using the extensive subtree-pruning-regrafting tree search algorithm.
Genomic tree inference: We applied three different genomic tree inference analyses: coalescent-based species tree inference (applied to the SNP dataset), maximum likelihood tree inference (applied to concatenated read supermatrix), and maximum parsimony tree inference (applied to the concatenated SNP dataset). We performed species tree inference using SNAPP (1 29) as implemented in BEAST 2.3.0, performing two iterations using default settings. Tracer 1.6 was used after each iteration to ensure that the runs had reached convergence, after which LogCombiner v2.3.0 and DensiTree (13 0) were used to process results. We carried out ML tree inference using RAxML v8.2.9 ( 96) on the concatenated read supermatrix using the general-time-reversible gamma-invariant (GTRGAMMA) model and performed 1,000 rapid bootstrap searches. Finally, we conducted conventional MP analysis on the concatenated SNPs of each taxon set, using the seqboot, dnapars, and consense algorithms in PHYLIP v3.695 (12 7) and running 10,000 bootstrap replicates to evaluate branch support.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |