Paraclevelandia brevis Kidder, 1937
|
publication ID |
https://doi.org/ 10.5852/ejt.2020.697 |
|
publication LSID |
lsid:zoobank.org:pub:8962B6E6-B278-4EF5-9E62-3E858726E2F2 |
|
DOI |
https://doi.org/10.5281/zenodo.4328999 |
|
persistent identifier |
https://treatment.plazi.org/id/44755610-B002-FF87-FDD4-FA60BAC07684 |
|
treatment provided by |
Valdenar |
|
scientific name |
Paraclevelandia brevis Kidder, 1937 |
| status |
|
Paraclevelandia brevis Kidder, 1937
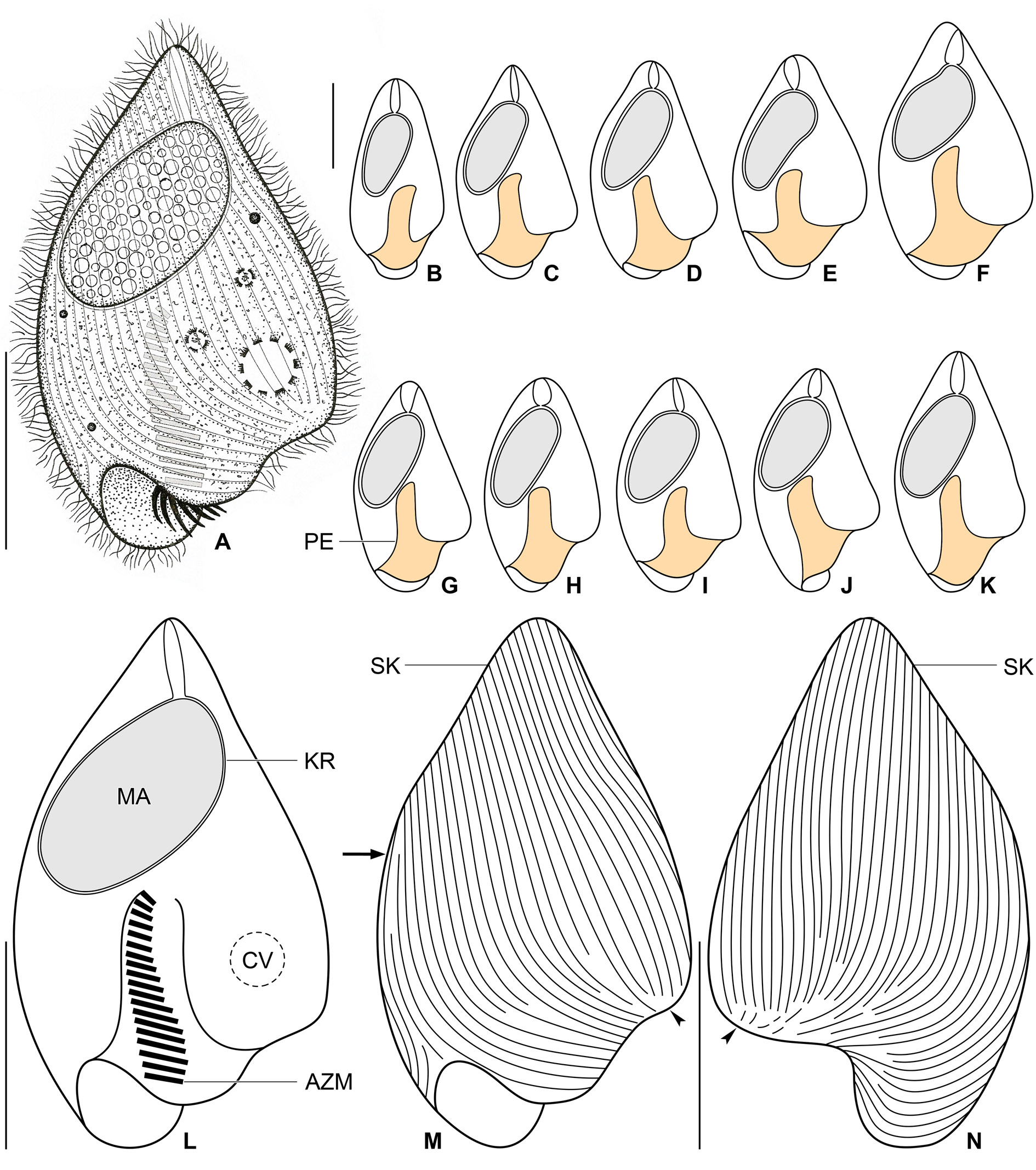
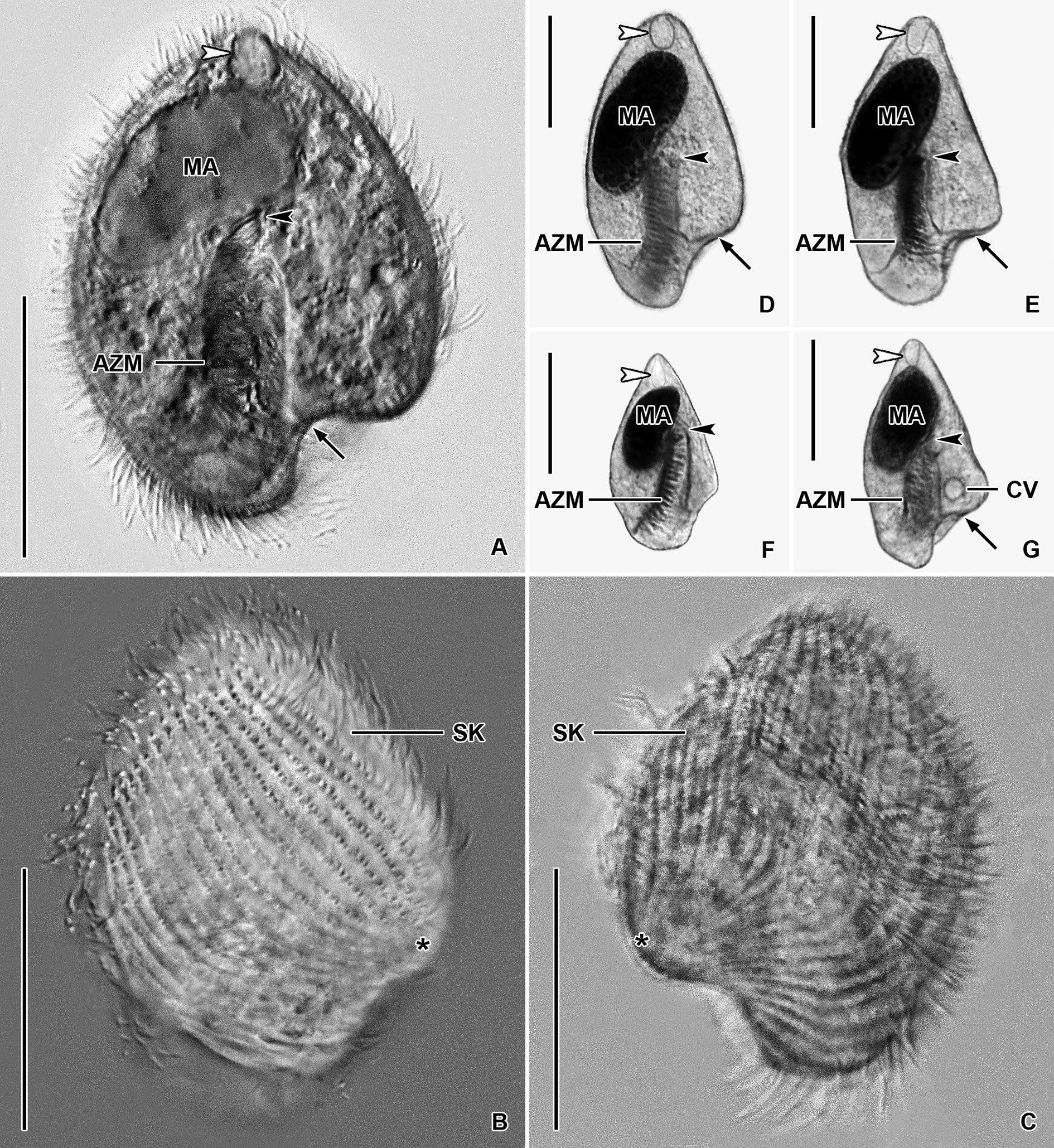
Figs 12–14 View Fig View Fig View Fig
Description of Vietnamese population
Size in vivo about 45–60 × 20–35 μm, usually 50 × 30 μm, as calculated from some in vivo measurements and morphometric data; length:width ratio ranging from 1.7:1 to 2.2:1 after protargol impregnation ( Table 5). Body obcordiform, widest at beginning of posterior third, i.e., about at level of contractile vacuole; dorsoventrally flattened 1.4:1. Anterior end usually bluntly pointed, rarely pointed; posterior body portion broadly rounded; left side slightly curved at level of peristomial opening and thus forming a more or less noticeable lobe; edge of dorsal side longer than that of ventral side and hence forming a distinct lip overhanging peristomial opening ( Figs 12 View Fig A–N, 14D–E). Macronucleus located in anterior half of body; ellipsoidal with a length:width ratio of 1.8–2.3: 1 in protargol preparations; 18–23 × 9–12 μm in size after protargol impregnation; filled with innumerable globular structures (very likely nucleoli) 0.7–1.9 μm in diameter after protargol impregnation, well observable in vivo and after protargol impregnation. Karyophore completely surrounds macronucleus, attaches to anterior body pole forming a distinct funnel ( Table 5; Figs 12 View Fig A–L, 14D–E). Micronucleus not observed. Contractile vacuole near left side at beginning of posterior body third, i.e., in indistinct lobe of left side ( Fig. 12A, L View Fig ). Cortex flexible, no cortical granules recognizable. Cytoplasm colorless; finely granulated; contains some free (symbiotic?) bacteria and/or archaea and food vacuoles about 2.6–3.7 μm across with prey prokaryotes ( Fig. 12A View Fig ). Swims slowly; dies quickly on microscope slides, possibly due to presence of oxygen.
Somatic ciliature holotrichous; cilia about 4.0–5.0 μm long in vivo and very narrowly arranged. Approximately 50 ciliary rows narrowly spaced over entire body surface. Almost all ciliary rows begin from a whorl (posterior suture) on left body side, near location of contractile vacuole, to radiate across ventral and dorsal sides toward right body margin; some kineties shortened posteriorly ( Fig. 12 View Fig M–N, arrowheads). Right suture extends from level of peristomial opening to anterior body end; formed by obliquely abutting ventral and dorsal ciliary rows ( Fig. 12M View Fig , arrow).
Peristomial opening situated on posterior body pole, occupies about 11% of body length and measures 4–6 × 10–17 μm after protargol impregnation ( Figs 12 View Fig A–M, 14D–E). Peristomial funnel about 17 μm long in protargol preparations. Adoral zone extends almost in parallel with main body axis along right side of peristomial funnel to terminate about at level of posterior end of macronucleus; occupies 34% to 41% of body length; composed of on average 18 membranelles; cilia of distalmost membranelles about 7 μm long in vivo and projecting out of peristomial funnel ( Table 5; Figs 12A, L View Fig , 14 View Fig D–E). Paroral membrane not observed.
Notes on Thai I population
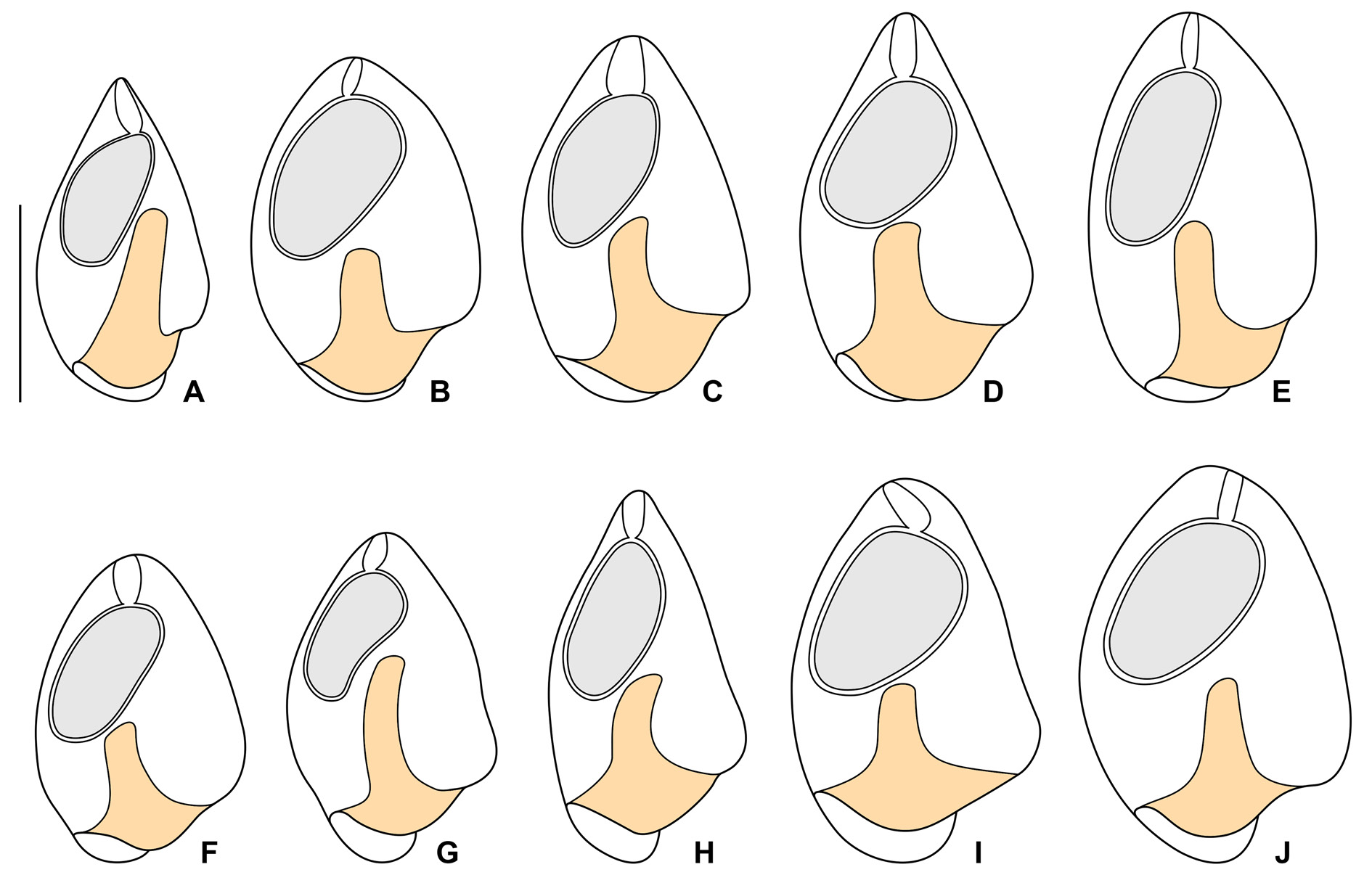
The Thai population matches very well the Vietnamese population. However, the body size of the Thai population is smaller than in the Vietnamese population (30–45 μm vs 45–60 μm). On the other hand, the peristomial funnel is longer in the Thai population than in the Vietnamese population (up to 55% vs 41% of body length). The variability of body size and shape of the Thai population is summarized in Table 5 and shown in Figs 13 View Fig A–J, 14A–C, F–G.
Morphometric and shape analyses
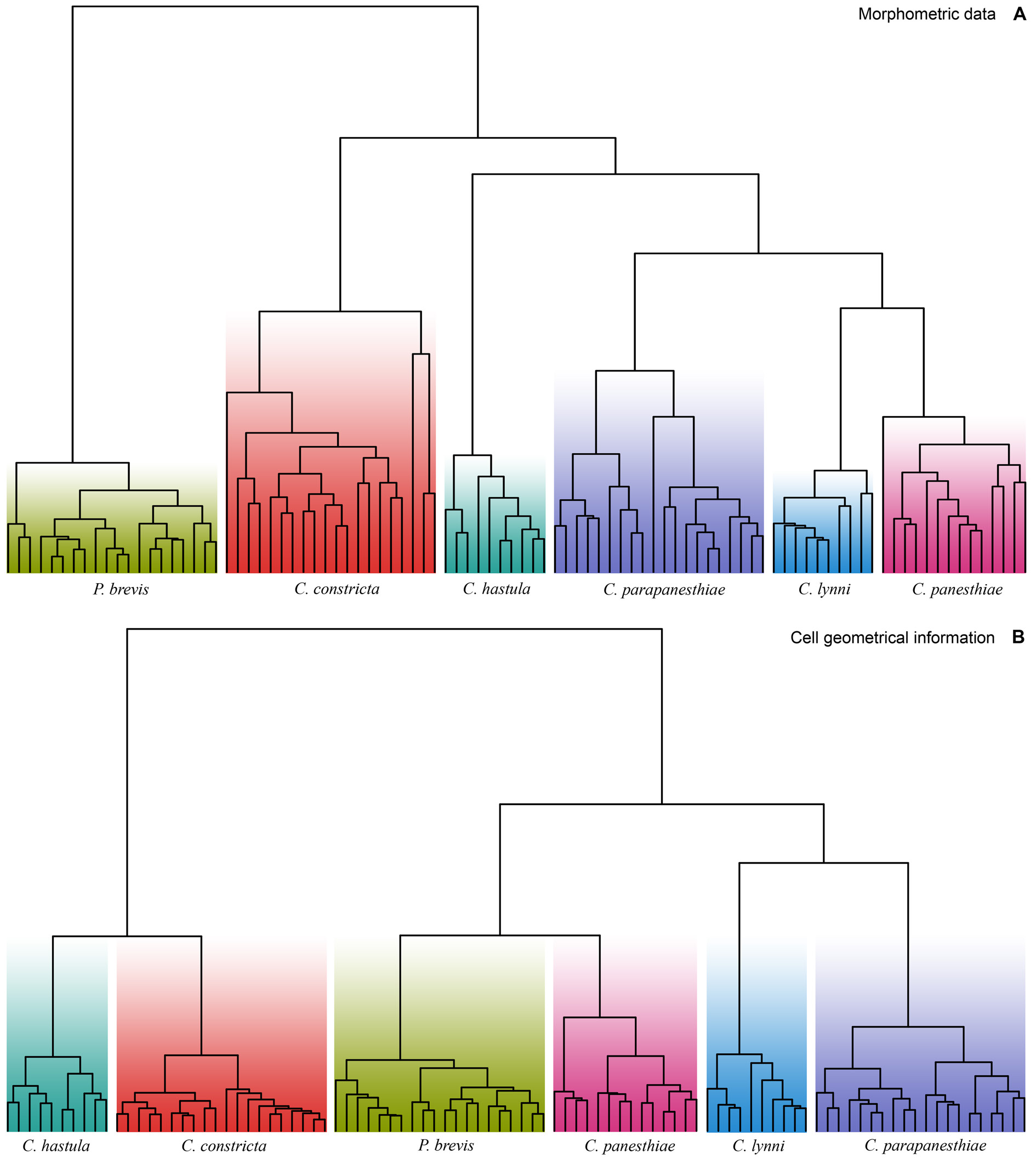
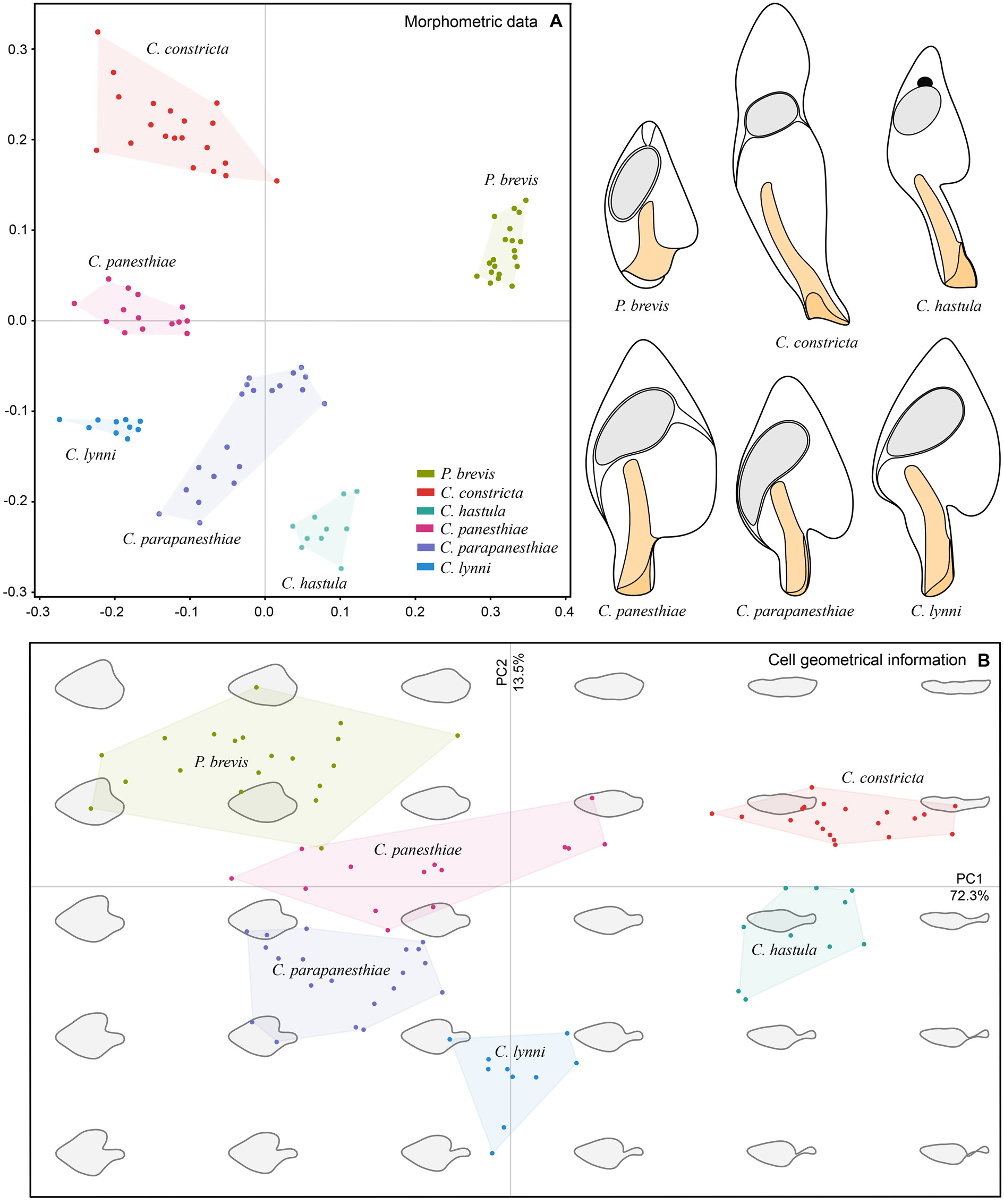
To assess the morphometric variation,distinctness,and boundaries of the six clevelandellid morphospecies, we utilized a multivariate approach including cluster analyses (CA) and metric multidimensional scaling (MDS). Altogether 16 morphometric features were used to calculate pairwise similarities (measured by Gower’s coefficient) of 20 Paraclevelandia and 74 Clevelandella specimens. The pairwise Gower’s similarity matrix was then subjected to six hierarchical CA, employing the average linkage, the weighted average linkage, the complete linkage, the median, the centroid, and the Ward’s clustering method. Since all clustering algorithms consistently depicted each morphospecies as a distinct group, we present here only one dendrogram that was produced by the weighted average linkage method ( Fig. 15A View Fig ). Likewise, MDS conducted on the Gower’s similarity matrix generated six mutually well-isolated and homogenous groups each representing one morphospecies ( Fig. 16A View Fig ).
Shape analyses, including CA and PCA, brought very similar results as did morphometric analyses, i.e., each morphospecies formed a well-delimited and homogenous group. We present here only one illustrative dendrogram that was computed with the Ward’s D2 clustering method in a combination with the Manhattan city block distance ( Fig. 15B View Fig ). This dendrogram was also completely consistent with the PCA diagram based on the Fourier coefficients ( Fig. 16B View Fig ). MANOVA performed on the PCA objects revealed statistically significant differences between shapes of individual clevelandellid morphospecies (Hotelling-Lawley trace = 48.25, approximate F 55, 382 = 67.03, P <2.2 × 10 –16).
Phylogenetic analyses
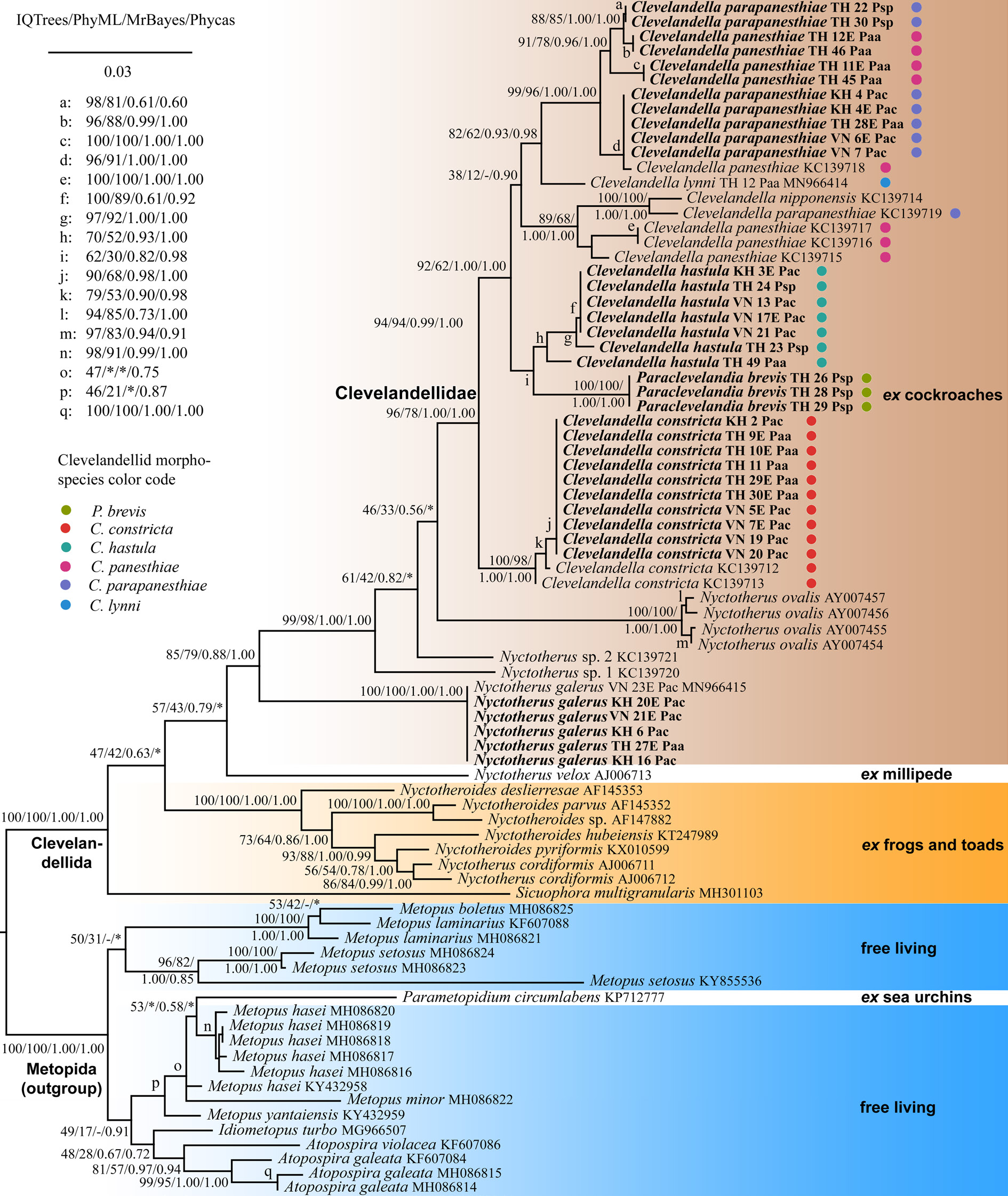
Thirty-six new 18S rRNA gene sequences of endozoic ciliates belonging to the order Clevelandellida , isolated from four populations of Panesthia cockroaches, were obtained. Length, GC content, and GenBank accession numbers of the new sequences are provided in Table 6. Four phylogenetic methods (IQTree, PhyML, MrBayes, and Phycas) were used to reconstruct relationships within the order Clevelandellida and to test for monophyly of the clevelandellid morphotypes. With respect to individual datasets, all phylogenetic methods resulted in similar topologies except for some statistically poorly supported nodes that might be considered as soft polytomies. Therefore, only IQTrees are presented along with nodal supports from all statistical methods ( Figs 17–18 View Fig View Fig ).
The larger dataset served, especially, to uncover the phylogenetic position of the family Clevelandellidae within the order Clevelandellida , to test for the monophyly of the family Clevelandellidae , and to reveal its fundamental bifurcation. Apart from the outgroup, the larger dataset included members of the family Sicuophoridae Amaro, 1972 (represented only by Sicuophora multigranularis Xiao et al., 2002 ), Nyctotheridae , and Clevelandellidae . In all analyses, S. multigranularis clustered with the Nyctotheridae + Clevelandellidae clade with full statistical support. The family Nyctotheridae was depicted as paraphyletic and contained the monophyletic family Clevelandellidae (96% IQTrees, 78% PhyML, 1.00 MrBayes, 1.00 Phycas). The Nyctotheridae + Clevelandellidae clade exhibited a clustering specific for higher taxa of their host organisms. One cluster received full statistical support in all analyses and comprised ciliates that had been isolated exclusively from the large intestine of amphibians ( Nyctotherus cordiformis (Ehrenberg, 1831) , Nyctotheroides deslierresae Affa’a, 1991, Nyctotheroides hubeiensis Li et al., 1998 , Nyctotheroides parvus (Walker, 1909) , Nyctotheroides pyriformis (Nie, 1932) , and Nyctotheroides sp. AF147882 View Materials ). The other cluster obtained poor statistical support (57% IQTrees, 43% PhyML, 0.79 MrBayes) and contained ciliates that had been isolated exclusively from the hindgut of arthropods (all remaining members of the family Nyctotheridae and all members of the family Clevelandellidae ). Within this weakly supported cluster, there was one very strongly supported node (99% IQTrees, 98% PhyML, 1.00 MrBayes, 1.00 Phycas), which united Nyctotherus sp. 1 KC139720 View Materials , Nyctotherus sp. 2 KC137721, four specimens of Nyctotherus ovalis Leidy, 1850 , and all clevelandellids. The newly sequenced specimens of Nyctotherus galerus Pecina & Vďačný, 2020 , which had been isolated from three Panesthia populations, were identical and grouped together with full statistical support. This species joined a cluster of all other ciliates isolated from cockroaches, forming a so-called ‘cockroach clade’ though with variable statistical support (85% IQTrees, 79% PhyML, 0.88 MrBayes, 1.00 Phycas). Nyctotherus velox Leidy, 1849 , isolated from a myriapod, was placed in a sister position to the ‘cockroach clade’ in most analyses. However, this grouping received only negligible statistical support (57% IQTrees, 43% PhyML, 0.79 MrBayes), and was not recognized in Phycas analyses at all. Therefore, we consider the phylogenetic position of N. velox to be questionable ( Fig. 17 View Fig ).
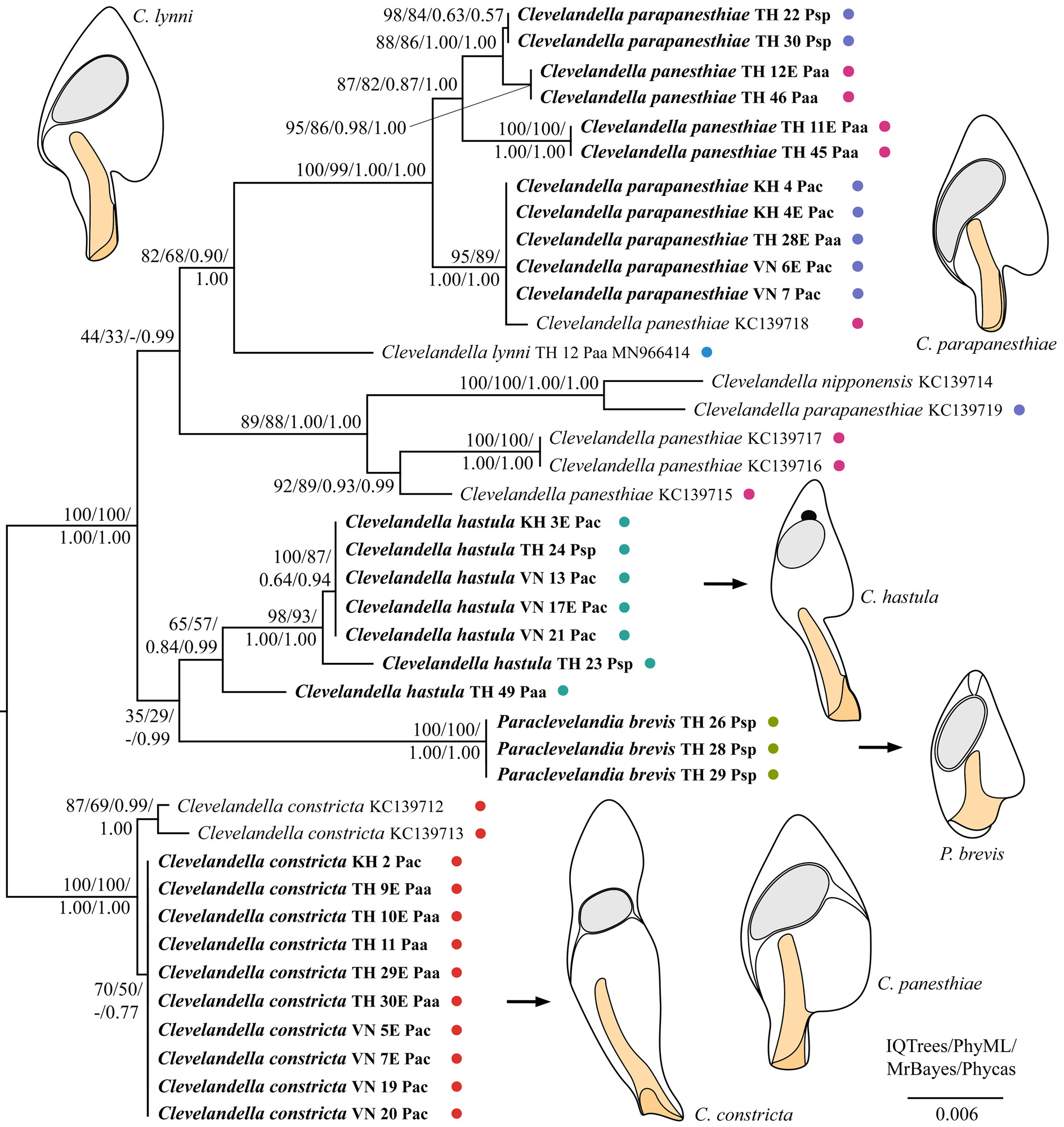
As concerns the family Clevelandellidae , its type genus Clevelandella was depicted paraphyletic because it contained members of the genus Paraclevelandia ( Figs 17–18 View Fig View Fig ). All sequences of P. brevis were identical and therefore grouped together with full statistical support in all analyses. Paraclevelandia brevis grouped with the C. hastula clade with variable support in both datasets. All members of the C. constricta morphotype clustered together with full or strong statistical support in all trees inferred from both datasets. Clevelandella constricta was depicted as sister to all other clevelandellids with strong statistical support. By contrast, the Asian and Australian specimens of the morphospecies C. panesthiae and C. parapaesthiae did not form a distinct cluster each, but were mixed together or grouped with other Clevelandella species. Namely, C. panesthiae specimens from the Thai I population were classified in two clusters, one of which grouped with C. parapanesthiae isolates from the Thai II population. The Australian C. panesthiae KC139718 View Materials specimen clustered together with C. parapanesthiae specimens from the Cambodian, Thai I, and Vietnamese populations. The 18S rRNA gene sequences of all these C. parapanesthiae specimens were identical and differed from the C. panesthiae KC139718 View Materials exemplar in only 2 nucleotide positions. The remaining Australian C. panesthiae specimens ( KC139715 View Materials , KC139716 View Materials , KC139717 View Materials ) formed a well-supported and structured clade, indicating that they belong to two distinct species. The Australian C. parapanesthiae KC139719 View Materials individual grouped with the Australian C. nipponensis KC139714 View Materials specimen with full statistical support in all analyses ( Figs 17–18 View Fig View Fig ).
To summarize, monophylies of only three morphospecies, viz., C. constricta , N. galerus and P. brevis , were strongly statistically supported. Nevertheless, we cannot exclude that the C. constricta morphotype comprises two distinct molecular species (genotypes), as suggested by the substructure within this morphotype in 18S rRNA gene trees inferred from the smaller dataset ( Fig. 18 View Fig ). According to the present phylogenetic analyses, the C. panesthiae morphotype unites at least five distinct molecular species, the C. parapanesthiae morphospecies contains at least three molecular species, and the C. hastula morphospecies covers at least three molecular species (genotypes).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |