Geocerthia Chesser and Claramunt
|
publication ID |
https://doi.org/ 10.5281/zenodo.189906 |
|
DOI |
https://doi.org/10.5281/zenodo.6220760 |
|
persistent identifier |
https://treatment.plazi.org/id/03F58782-FF91-4671-FF24-FAE1FC2AE2A9 |
|
treatment provided by |
Plazi |
|
scientific name |
Geocerthia Chesser and Claramunt |
| status |
|
Geocerthia Chesser and Claramunt , genus nov.
Type species. Upucerthia serrana Taczanowski, 1875 .
Included species. Geocerthia serrana ( Taczanowski, 1875) comb. nov., Striated Earthcreeper.
Diagnosis, morphology. Large earthcreeper (19–20 cm, 44–52 g, Remsen 2003). Bill longish, decurved; face grizzled brown and whitish; whitish superciliary; crown medium-dark brown with pale streaking, especially on forehead; back medium brown with faint pale streaking; wings, tail, and uppertail coverts rufous; throat whitish; underparts dull gray-brown with prominent pale streaking. Geocerthia differs from true Upucerthia earthcreepers, which have long, thin, highly decurved bills, by its comparatively shorter and stouter decurved bill and its overall darker plumage. Distinguished from all Cinclodes and Upucerthia species by its rufous wings, uppertail coverts, and tail. Lacks the wingband typical of Cinclodes species.
Etymology. From the Greek geo (earth) and certhia (treecreeper), referring to the terrestrial habits of G. serrana and to its bill, which resembles those of the treecreepers. The construction of the name is a direct parallel to the English name earthcreeper. The name is feminine.
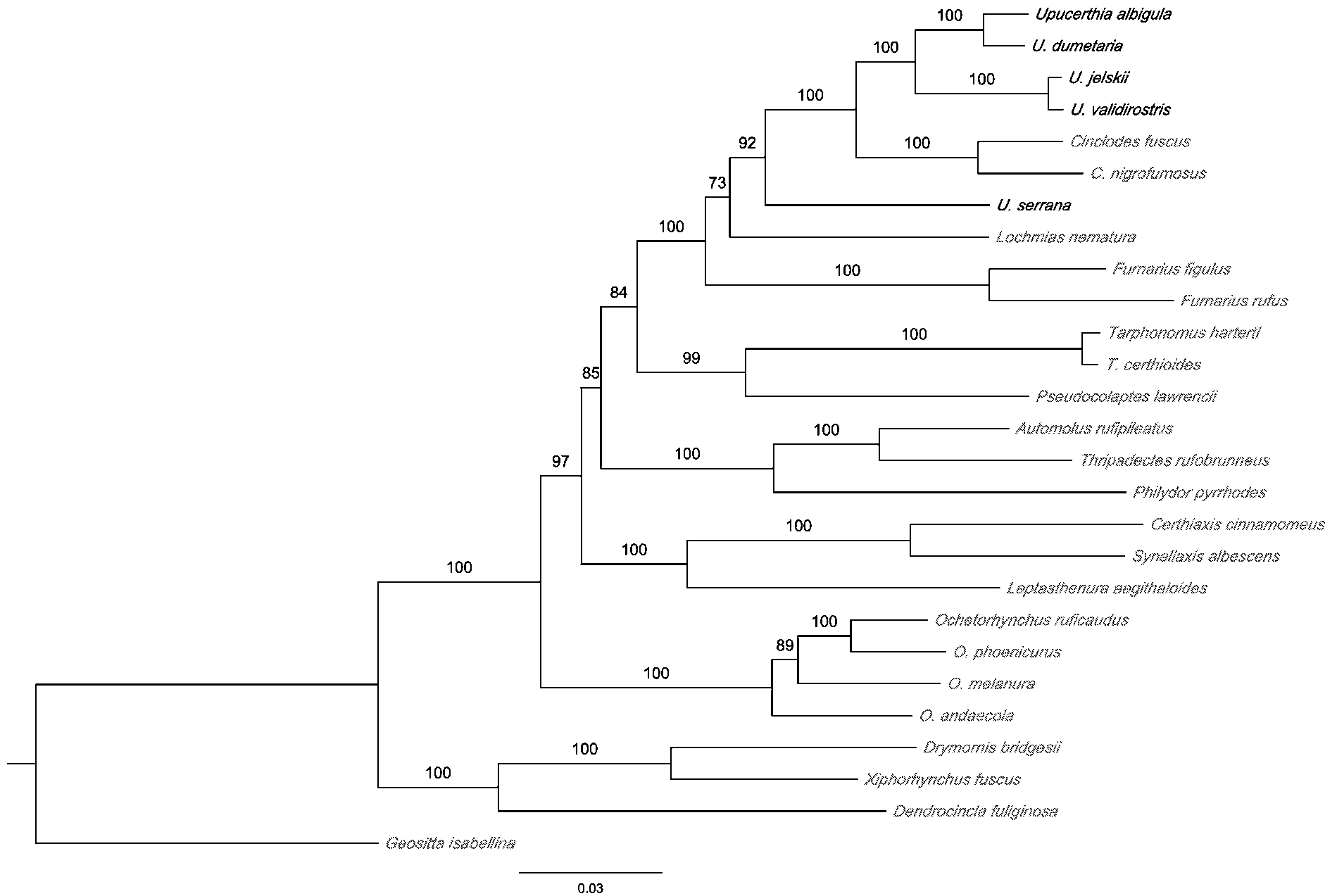
Molecular analyses. A molecular analysis of furnariid species revealed that Geocerthia is sister to a clade composed of all species of Cinclodes and all species of Upucerthia sensu stricto, which in turn are sister taxa. To demonstrate that Geocerthia and Upucerthia are not sister genera, we present an analysis of a subset of taxa from the larger study. Taxon sampling for this subset (Table 1) included representatives of U. serrana , all other species of the traditional genus Upucerthia ( Upucerthia , Tarphonomus , Ochetorhynchus ), two species of Cinclodes , and single species of the genera Furnarius , Leptasthenura , Synallaxis , Certhiaxis , Pseudocolaptes , Philydor , Thripadectes , Automolus , and Lochmias . We used as outgroups the dendrocolaptines Dendrocincla fuliginosa , Drymornis bridgesii , and Xiphorhynchus fuscus , as well as the basal furnariid Geositta isabellina . As a simple test of its monophyly and phylogenetic relationships, a second individual of Geocerthia serrana (ZMUC [Zoological Museum, University of Copenhagen] S444; GenBank Accession #s: EF635339 View Materials , EF635358 View Materials , EF635319 View Materials ) was sequenced. This individual differed from the first by only one nucleotide substitution and grouped with it in all analyses in which it was included (results not shown).
Table 1. Tissues used in the phylogenetic analysis.
Taxon Museum Tissue ID Locality
Geositta isabellina AMNH 12181 CHILE: Region Metropolitana, 15 km ENE Embalse El Yeso, ca. 3400m
Upucerthia dumetaria AMNH 10396 ARGENTINA: Prov. Neuquén, Sierra Auca Mahuida
. albigula LSUMNS B61491 PERU: Depto. Arequipa, 14 km E Pancarpata, 3222m
. jelskii LSUMNS B103886 PERU: Depto. Tacna, Tacna-Ilave Road, ca 25 km NE Tarata, 4050m. validirostris LSUMNS B17160 ARGENTINA: Prov. Tucumán, 12 km N, 4 km W Tafi de Valle, 3060 m. serrana LSUMNS B49662 PERU: Depto. Lima, Santa Eulalia road, ca 86 km NE Lima Tarphonomus certhioides LSUMNS B18872 BOLIVIA: Depto. Santa Cruz, Estancia Perforación, ca 130 km E Charagua, 520
m
harterti LSUMNS B34573 BOLIVIA: Depto. Santa Cruz, Tambo, 14 km SE Camarapa Ochetorhynchus andaecola LSUMNS B1199 BOLIVIA: Depto. La Paz;, 2.5 km by road S Mecapaca, ca 26 km by road S
Calacoto, 3050m
. ruficaudus LSUMNS B103908 PERU: Depto. Arequipa, km 60 on Div. Arequipa-Juliaca road, ca 10 road km W
Chaguata
. phoenicurus AMNH 9943 ARGENTINA: Prov. Río Negro, 20 km E Ñorquinco, 1000 m. melanura AMNH 12148 CHILE: Region Metropolitana, ca 4 km SSW by road from peak of Cerro de El
Roble, ca 1600 m
Cinclodes fuscus AMNH 12169 CHILE: Region Metropolitana, 2 km ENE Embalse El Yeso, ca. 2500 m Taxon Museum Tissue ID Locality
. nigrofumosus AMNH 12164 CHILE: Region Valparaíso, Roca Brava, ca. 2 km N Zapallar Furnarius figulus FMNH 392828 BRAZIL: Estado Alagoas, Piranhas, Fazenda Bela Vista
. rufus AMNH 10431 ARGENTINA: Prov. Neuquén, Centenario
Leptasthenura aegithaloides AMNH 10306 ARGENTINA: Prov. Neuquén, Sierra Auca Mahuida
Synallaxis albescens AMNH 2295 BOLIVIA: Depto. Santa Cruz, 300 m N of Rio Mercedes, 600 m Certhiaxis cinnamomeus AMNH 6190 BOLIVIA: Depto. Santa Cruz, 300 m N of Rio Mercedes, 600 m Pseudocolaptes lawrencii AMNH 3694 COSTA RICA: Prov. Herodia, 3 km N Porrosati, 2200 m Philydor pyrrhodes AMNH 8864 VENEZUELA: Estado Amazonas, Mrakapiwie
Thripadectes rufobrunneus AMNH 3651 COSTA RICA: Prov. San Jose, Cerro de la Muerte, 3350 m Automolus rufipileatus LSUMNS B1074 BOLIVIA: Depto. La Paz, Río Beni, ca 20 km by river N Puerto Linares, 600 m Lochmias nematura AMNH 12074 ARGENTINA: Prov. Misiones, Parque Provincal Urugua-i, ca. 1 km W park
headquarters, Ruta Provincial 19, ca 400 m
Dendrocincla fuliginosa AMNH 12706 VENEZUELA: Estado Amazonas; Río Baria, Cerro de la Neblina Base Camp Drymornis bridgesii LSUMNS B25799 PARAGUAY: Depto. Alto Paraguay, Madrejón
Xiphorhynchus fuscus LSUMNS B35576 BRAZIL: Estado Bahia, ca 16 km W Porto Seguro RPPN Vera Cruz Tissue collections: LSUMNS – Louisiana State University Museum of Natural Science, Baton Rouge; AMNH – American Museum of
Natural History, New York; FMNH – Field Museum of Natural History, Chicago.
Total DNA was extracted from 25 mg of pectoral muscle using the DNeasy kit (Qiagen) and the standard protocol. Following methods described in Chesser et al. (2007), we amplified and sequenced the mitochondrial genes ND3 and CO2, and the autosomal nuclear gene beta-fibrinogen intron 7. Following the same methods, we amplified and sequenced an additional mitochondrial gene (ND2) using the primers H6313 ( Johnson and Sorenson 1998) and L5215 ( Hackett 1996). Two additional nuclear protein-coding genes (RAG1 and RAG2) were included for one individual from each genus. These RAG sequences were taken from Moyle et al. (2009) except for those of Geocerthia serrana , which we amplified and sequenced following the methods in Moyle et al. (2009). The six-gene concatenated dataset included 6,970 base pairs after alignment issues and unique inserts were excluded from BF7, the nuclear intron. An exception to a complete dataset was a partial BF7 sequence for the individual representing Ochetorhynchus andaecola . In addition, the ND2 gene could not be amplified for the selected individuals of Philydor pyrrhodes or Lochmias nematura ; ND2 sequence from a second individual of these same species (amplified for a separate analysis) was used instead.
Because the dataset included protein-coding mitochondrial genes, a nuclear intron and protein-coding nuclear genes, it was unlikely that a single nucleotide substitution model would provide the best fit to the data. To determine the best partition of the dataset, we evaluated six different partitioning regimes ranging from unpartitioned to the maximum of sixteen partitions. Partitions were examined by performing maximum likelihood (ML) analyses using RAxML 7.0.4 on the Cipres Portal V 1.5 (http://www.phylo.org/sub_sections/portal/). RAxML implements the GTR+Γ model and this was applied in each partition regime. We used the Akaike Information Criterion (AIC) to choose the best partitioning strategy, which was the GTR+ Γ+I model and fully partitioned dataset. We then used RAxML to evaluate nodal support by performing 150 bootstrap replicates ( Stamatakis et al. 2008). The analysis resulted in a single maximum-likelihood tree (-log L = 28529.0) with high bootstrap support for most relationships ( Fig. 1 View FIGURE 1. A ). Individuals of Upucerthia sensu stricto formed a strongly supported clade (100% bootstrap) that was sister to Cinclodes . The Upucerthia - Cinclodes clade was well supported (100%), and the single species G. serrana received strong support as sister to this clade (92%). These results clearly demonstrate that G. serrana is not a member of the Upucerthia clade.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.