Neoechinorhynchus (Neoechinorhynchus) schmidti Barger, Thatcher & Nickol, 2004
|
publication ID |
https://doi.org/ 10.11646/zootaxa.3985.1.5 |
|
publication LSID |
lsid:zoobank.org:pub:E6FF2FAB-4C99-4E4A-8E56-8D9B5A161474 |
|
DOI |
https://doi.org/10.5281/zenodo.5685263 |
|
persistent identifier |
https://treatment.plazi.org/id/03919446-652F-434E-8CE0-F8FFFEA2C3A7 |
|
treatment provided by |
Plazi |
|
scientific name |
Neoechinorhynchus (Neoechinorhynchus) schmidti Barger, Thatcher & Nickol, 2004 |
| status |
|
Neoechinorhynchus (Neoechinorhynchus) schmidti Barger, Thatcher & Nickol, 2004 View in CoL
( Fig. 2 View FIGURE 2 i)
Mexico: TABASCO: 24 km to S of Villahermosa; Trachemys venusta (Barger 2004; Barger et al. 2004). Pantanos de Centla (18°28’18.9’’N 92°39’14.9’’W). Trachemys scripta ( Martínez-Aquino et al. 2009; García-Varela et al. 2011).
Specimens deposited. CNHE (6764), HWML (17667), USNPC (92883).
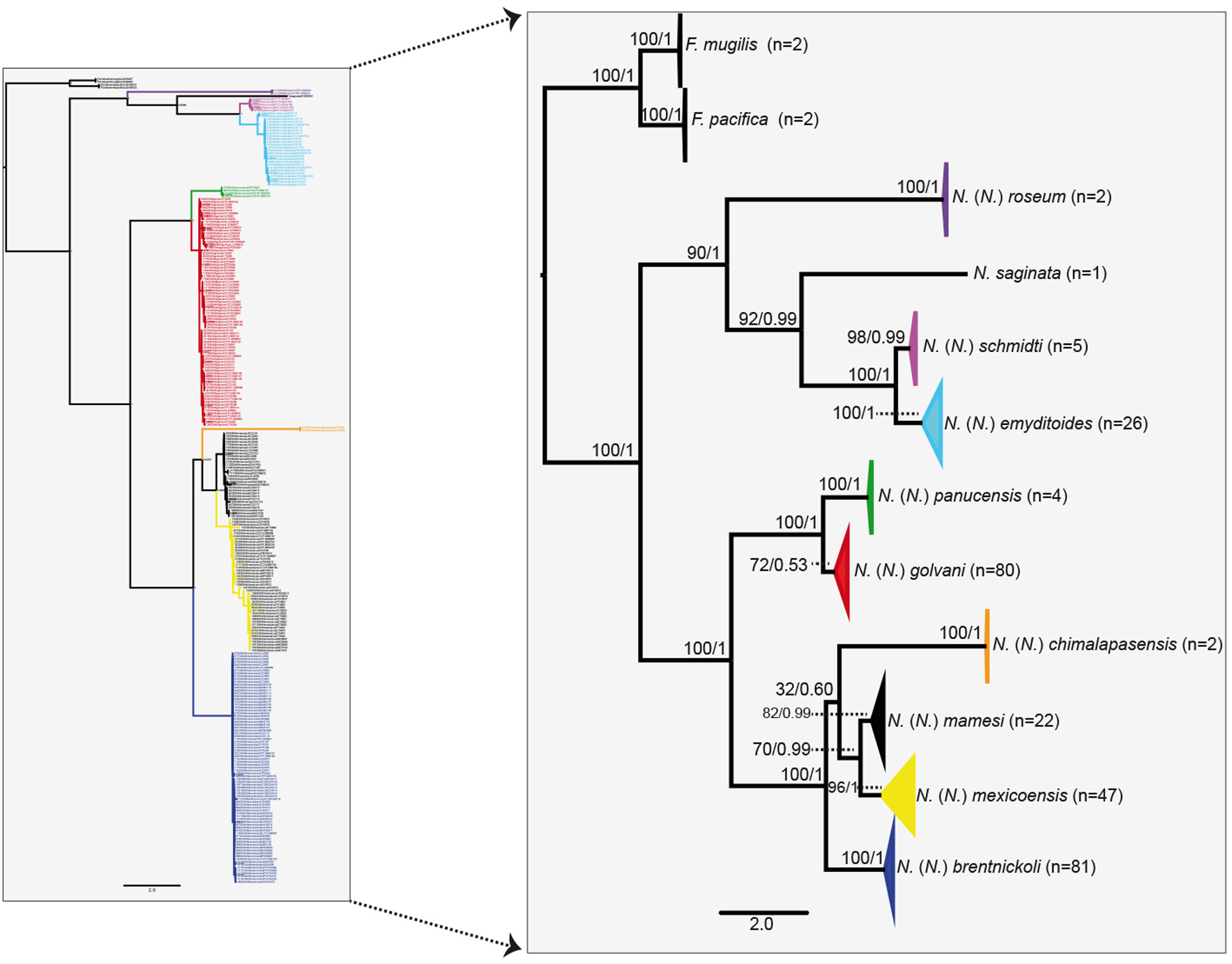
Species delimitation through phylogenetic analysis and genetic divergence. To demonstrate the species boundaries among the species reported in this checklist, we sequenced a total of 269 individuals of the species: N. (N.) brentnickoli (81 individuals from 16 localities); N. (N.) roseum (two individuals from two localities); N. (N.) mamesi (22 individuals from seven localities); N. (N.) golvani (80 individuals from 11 localities); N. (N.) mexicoensis (47 individuals from eight localities); N. (N.) schmidti (five individuals from one locality); N. (N.) emyditoides (26 individuals from three localities); N. (N.) panucensis (four individuals from two localities); and N. (N.) chimalapasensis (two individuals from one locality). Additionally sequence of N. saginata plus Floridosentis mugilis and Floridosentis pacifica were aligned together, composing a data set of 806 bp with 274 terminals. Nucleotide frequencies for 28S rDNA data set were 0.246 (A), 0.178 (C), 0.276 (G), and 0.298 (T). Phylogenetic analyses of the available sequences for the genus Neoechinorhynchus through the Bayesian consensus tree yielded the same branch pattern as the ML tree ( Fig. 3 View FIGURE 3 ). Both trees yielded that the ten species of Neoechinorhynchus are monophyletic with strong nodal support ranging from 72 to 100 % of bootstrap, and 1.0 of posterior probability ( Fig. 3 View FIGURE 3 ). Both trees yielded three major clades. The first clade contained four species N. (N.) roseum , N. saginata , N. (N.) schmidti and N. (N.) emyditoides . The second clade was composed of two species N. (N.) golvani and N. (N.) panucensis and had 100 % bootstrap support and 1.0 posterior probability. The third clade was composed of four species N. (N.) chimalapasensis , N. (N.) brentnickoli , N. (N.) mamesi and N. (N.) mexicoensis and received reliable bootstrap support and high values of posterior probability ( Fig. 3 View FIGURE 3 ).
67- 76 N. (N.) brentnickoli 10 D. latifrons Laguna de Tres Palos, Guerrero, México FJ388991 View Materials , 8178 FJ968156 View Materials - FJ968159 View Materials , KR086219 View Materials - KR086223 View Materials *
77- 81 N. (N.) brentnickoli 5 D. latifrons Río Tamarindo, Chautengo , Guerrero, KR086224 View Materials - KR086228 View Materials * 8179 México
82 N. (N.) roseum 1 Achirus mazatlanus Laguna el Caimanero, Sinaloa, México FJ388999 View Materials 6762, 633, 634 83 N. (N.) roseum 1 Citharichthys gilberti Estero La Tovara , Nayarit, México FJ389000 View Materials 6763 84- 86 N. (N.) mamesi 3 D. latifrons Puente Manialtepec , Oaxaca KR086229 View Materials - KR086231 View Materials *
......continued on the next page TABLE 1 View TABLE 1 . (Continued)
Samples Species N Host Locality GenBank Specimensdeposited
(DNA) CNHE
87- 91 N. (N.) mamesi 5 D. latifrons Estero Joaquin Amaro , Chiapas, México JN830770 View Materials - JN830774 View Materials 8183
92- 95 N. (N.) mamesi 4 D. latifrons Laguna la Conquista, Chiapas, México JN830766 View Materials - JN830769 View Materials 8184
96- 98 N. (N.) mamesi 3 D. latifrons Laguna Rion Pijijiapan , Chiapas, JN830763 View Materials - JN830765 View Materials 8180, 8181, 8182
México
99- 100 N. (N.) mamesi 2 D. latifrons Quebrada Ganados , Costa Rica KR086232 View Materials *, KR086233 View Materials * 8590
101- 102 N. (N.) mamesi 2 D. maculatus Playa Grande , Costa Rica KR086234 View Materials *, 8191 KR086235 View Materials *
103- 105 N. (N.) mamesi 3 D. latifrons Las lisas, Guatemala KR086236 View Materials - KR086238 View Materials *
106- 107 N. (N.) golvani 1 Parachromis Lago Jalapa, Costa Rica KR086239 View Materials *
108- 109 N. (N.) golvani 2 Parachromis loisellei Lago Jalapa, Costa Rica KR086240 View Materials *,
KR086241 View Materials *
110- 111 N. (N.) golvani 2 Amphilophus Lago Jalapa, Costa Rica KR086242 View Materials *, KR086243 View Materials * 8592 longimanus
112 N. (N.) golvani 1 Heterotilapia Lago Jalapa, Costa Rica KR086244 View Materials *
113- 114 N. (N.) golvani 2 Archocentrus Lago Jalapa, Costa Rica KR086245 View Materials *,
centrarchus KR086246 View Materials *
115- 116 N. (N.) golvani 2 Amatitlania Quebrada Puercos , Costa Rica FJ388998 View Materials , 6757 nigrofasciata KR086247 View Materials *
117- 127 N. (N.) golvani 11 Vieja pearsei Presa Chicoasen , Chiapas, México FJ388995 View Materials , FJ968136 View Materials - FJ968138 View Materials , 6755 KR086248 View Materials - KR086254 View Materials *
128- 132 N. (N.) golvani 5 V. pearsei Presa Nezahualcoyolt, Chiapas, México FJ388996 View Materials , FJ968141 View Materials , FJ968142 View Materials , 6756 KR086255 View Materials *,
KR086256 View Materials *
133- 143 N. (N.) golvani 11 Parachromis Lago Canitzan, Tenosique , Tabasco, FJ388994 View Materials , FJ968139 View Materials , FJ968140 View Materials , 6767 friedrichstalii México KR086257 View Materials - KR086264 View Materials *
144- 153 N. (N.) golvani 10 Cichlasoma Río Carrizal , Tabasco, México FJ388993 View Materials , FJ968134 View Materials , FJ968135 View Materials , 6754 urophthalmum KR086265 View Materials - KR086271 View Materials *
154- 156 N. (N.) golvani 3 C. urophthalmum Lago las Ilusiones, Tabasco, México FJ388992 View Materials - FJ968144 View Materials
157- 167 N. (N.) golvani 11 Paraneetroplus Lago de Catemaco, Veracruz, México FJ388986 View Materials , FJ968145 View Materials , FJ968146 View Materials , 601, 603, 604, 606, 631,
fenestratus KR086272 View Materials - KR086279 View Materials * 632, 6783
168- 172 N. (N.) golvani 5 C. urophthalmum Presa Temascal , Oaxaca, México KR086280 View Materials - KR086284 View Materials * 8593
......continued on the next page TABLE 1 View TABLE 1 . (Continued) The sequence divergence estimated among the nine species herein considered, plus an additional sequence available for a species that occurs in freshwater fishes in North America ( N. saginata ) ranged from 3.2% to 48.7% ( Table 2). Instead, the intraspecific genetic divergence varied markedly among all the isolates sequenced for each acanthocephalan species and it was never higher than 3% for all intraspecific comparisons. For instance, the divergence among 81 specimens of N. (N.) brentnickoli obtained from a wide geographic range, in 16 localities (localities 1-16 in Fig. 1 View FIGURE 1 ) across the Pacific coast of northwestern Mexico, varied from 0 to 0.7% ( Table 2). In other example, N. (N.) roseum corresponding to two individuals of two localities 1) Laguna el Caimanero, Sinaloa and 2) Estero La Tovara, Nayarit (localities 17-18), had genetic divergence from 0 to 0.04%. The species N. (N.) mamesi comprises 22 individuals from seven localities from Mexico, one of Guatemala and two from Costa Rica (localities 19-25 in Fig. 1 View FIGURE 1 ), showing a genetic divergence from 0 to 1.3 %. N. (N.) golvani shows genetic divergence from 0 to 1.5% among 80 specimens from 11 localities (see localities 26-36). The species N. (N.) mexicoensis corresponding to 47 individuals from eight localities (localities 37-44) from Gulf of Mexico shows a genetic divergence ranged from 0 to 3%. N. (N.) schmidti shows low genetic divergence ranged from 0-0.02% of specimens analyzed from the same locality. The species N. (N.) emyditoides corresponding three localities: 1) Lago de Catemaco, Veracruz, 2) Río Papaloapan, Tlacotalpan, Veracruz and 3) Presa la Herradura, Monterrey (localities 46-48), showed a genetic divergence from 0 to1.3%. The specimens of N. (N.) panucensis are from Axtlan de Terrazas, San Luis Potosí, and Río Pantepec, Veracruz (localities 49-50), both localities are near of the type locality of N. (N.) panucensis . The genetic divergence among four isolates of N. (N.) panucensis ranged from 0.02–0.05%. Finally, the two specimens of N. (N.) chimalapasensis showed a genetic divergence equal to zero (locality 51).
1 2 3 4 5
1. N. (N.) brentnickoli (n = 81) 0–0.7 42–43 8.1–9.8 21.9–23.7 7.3–10 2. N. (N.) roseum (n = 2) 0–0.04 40.9–42.9 41.1–43.1 39.7–43.2 3. N. (N.) mamesi (n = 22) 0–1.3 21–23.6 2.40–5.50 4. N. (N.) golvani (n = 80) 0–1.5 21.0–24.6 5. N. (N.) mexicoensis (n = 47) 0–3.0 6. N. (N.) schmidti (n = 5)
7. N. (N.) emyditoides (n = 26)
8. N. (N.) panucensis (n = 4)
9. N. (N.) chimalapasensis (n = 2)
10. N. saginata (n =1)
continued.
TABLE 1. Specimens information: collection sites (CS), samples analyzed for DNA, species analyzed, specimens examined (N), host species, localities, GenBank accession number and catalog number (CNHE) for specimens studied in this work. Sequences marked with an asterisk (*) were obtained in the current study.
| Samples (DNA) 1-3 4-8 9-12 | Species N. (N.) brentnickoli N. (N.) brentnickoli N. (N.) brentnickoli | N Host 3 Dormitator latifrons 5 D. latifrons 4 D. latifrons | Locality Todos los Santos, Baja California Sur, México Oasis San José del Cabo, Baja California Sur, México Topolobampo,Sinaloa,México | GenBank KR086167 View Materials - KR086169 View Materials * KR086170 View Materials - KR086174 View Materials * KR086175 View Materials - KR086178 View Materials * | Specimensdeposited CNHE 8586 |
|---|---|---|---|---|---|
| 13-17 | N. (N.) brentnickoli | 5 D. latifrons | El Huizache, Sinaloa, México | KR086179 View Materials - KR086183 View Materials * | 8587 |
| 18-22 | N. (N.) brentnickoli | 5 D. latifrons | Vía Ferrea, Escuinapa, Sinaloa, México | KR086184 View Materials - KR086188 View Materials * | 8588 |
| 23-27 | N. (N.) brentnickoli | 5 D. latifrons | Nuevo Vallarta, Nayarit, México | KR086189 View Materials - KR086193 View Materials * | |
| 28-33 34 | N. (N.) brentnickoli N. (N.) brentnickoli | 6 D. latifrons 1 D. latifrons | Estero Chamela, Jalisco, México Río Cuitzmala, Jalisco, México | FJ388990 View Materials , KR086194 View Materials - KR086198 View Materials * FJ388989 View Materials | 8589 |
| 35-39 | N. (N.) brentnickoli | 5 D. latifrons | Laguna de Cuyutlan, Colima, México | KR086199 View Materials - KR086203 View Materials * | |
| 40-49 50-54 | N. (N.) brentnickoli N. (N.) brentnickoli | 10 D. latifrons 5 D. latifrons | Estero Boca de Apiza, Michoacán, México Estero Huahua, Michoacán, México | JN830782 View Materials - JN830786 View Materials , KR086204 View Materials - KR086208 View Materials * KR086209 View Materials - KR086213 View Materials * | 8176 8177 |
| 55-59 60-64 65-66 | N. (N.) brentnickoli N. (N.) brentnickoli N. (N.) brentnickoli | 5 D. latifrons 5 D. latifrons 2 D. latifrons | Estero Mexcalhuacan, Michoacán, México Estero Barra de Pichi, Michoacán, México Laguna de Coyuca, Guerrero, México | JN830777 View Materials - JN830781 View Materials KR086214 View Materials - KR086218 View Materials * JN830775 View Materials , JN830776 View Materials | 8173 8174 8175 |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |