Steganacarus ( Steganacarus ) similis Willmann, 1939
|
publication ID |
https://doi.org/10.11646/zootaxa.3664.4.9 |
|
publication LSID |
lsid:zoobank.org:pub:8F3BC3F8-AE22-4570-92CC-5F4750090C5C |
|
DOI |
https://doi.org/10.5281/zenodo.5658129 |
|
persistent identifier |
https://treatment.plazi.org/id/EE6A87DC-FFA7-FFF8-46AE-F976FA8835C1 |
|
treatment provided by |
Plazi |
|
scientific name |
Steganacarus ( Steganacarus ) similis Willmann, 1939 |
| status |
|
Steganacarus ( Steganacarus) similis Willmann, 1939 View in CoL
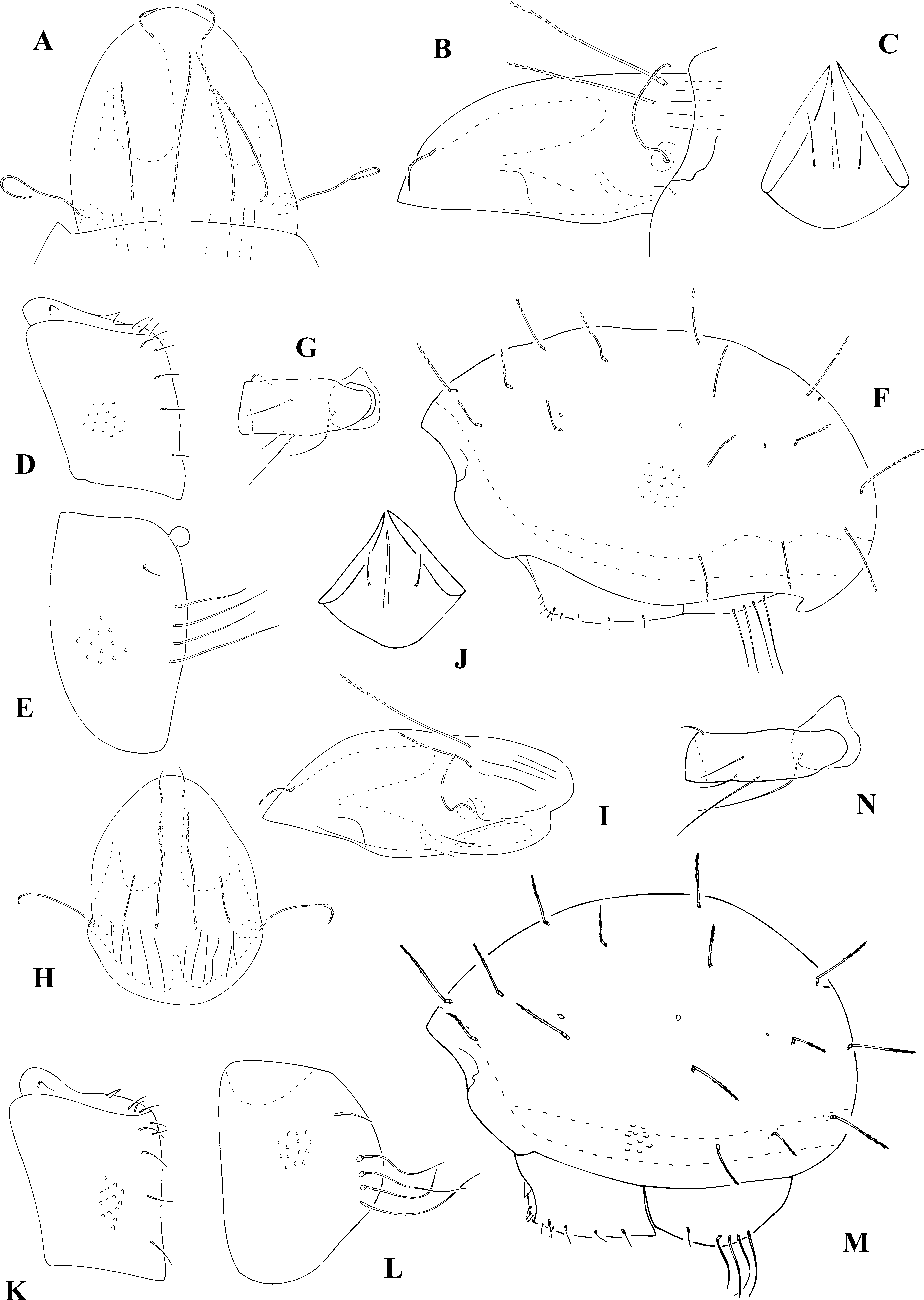
( Fig. 3A – G View FIGURE 3 A – N. A – G )
Steganacarus similis: Bernini and Magari 1993
Measurements of specimen from Rabaçal, Madeira. Prodorsum: length 257, height 111, width 202; setae of prodorsum: sensillum 96, interlamellar 139, lamellar 101, rostral 45, exobothridial 20; notogaster: length 596, height 353, width 364, setae of notogaster: c 1 88, c1 / c1 – d1 =0.7, h 1 and p 1 101; genitoaggenital plates: 156 × 106, anaoadanal plates: 207 × 116.
Diagnosis. Surface covered with feeble concavities. Colour brown. Prodorsum: median field narrow and longer than the laterals; lateral carinae absent; sensilla long, narrow, sickle-shaped covered with spines; setae, other than the exobothridial covered with distinct spines in distal half, rostral setae directed inward. Notogaster with rigid, short ( c1<c1 – d1) setae covered with spines in distal half; setae c1 and c3 near the anterior margin, setae c2 far away; vestigial setae f1 slightly posteriorly of setae h1; two pairs of lyrifissures ia and im. Ventral region: h > h – h; formula of genital setae: 6(4+2): 3, anoadanal plate with fairly long setae, ad1 and ad2 are longest, an / not =0.3. Chaetome of legs reduced; setae s on tarsi I are missing; v ”/ v’ =4.4.
Comparison. This species is very similar to the Paneuropean species S. ( S.) spinosus ( Fig. 3H – N View FIGURE 3 A – N. A – G ). The comparison was made with the specimens from the population collected in Poland (Sudety, Wielka Sowa).
Measurements of specimen of S. ( S.) spinosus (Sellnick, 1920) from Wielka Sowa, Poland. Prodorsum: length 303, height 126, width 212; setae of prodorsum: sensillum 94, interlamellar 152, lamellar 63, rostral 43, exobothridial 40; notogaster: length 606, height 404, width 434, setae of notogaster: c 1 121, c1 / c1 – d1 =0.66, h 1 and p 1 101; genitoaggenital plates: 167 × 126, anaoadanal plates: 202 × 131.
Steganacarus ( S.) similis differs from S. ( S.) spinosus also by two main morphological features. S. ( S.) similis from Madeira has longer lamellar setae ( le / in > 0.7), S. ( S.) spinosus has much shorter lamellar setae ( le / in <0.5); rostral setae in S. ( S.) similis are diriged inward but rostral setae of S. ( S.) spinosus are diriged forward.
Willmann (1939) differentiated S. ( S.) similis from S. ( S.) spinosus – see also Willmann (1931), but in the comparison he has put too much emphasis on the body size and he was incorrect in claiming that the setae of S. ( S.) spinosus are smooth and pointed distally.
Bernini and Magari (1993) have determined the neotype of S. ( S.) similis . They have pointed to some small morphological differences between S. ( S.) similis with S. ( S.) spinosus : in the shape of sensilla, shape of anoadanal setae, lack of setae a on the first tarsi, the length of anoadanal setae decreases gradually from ad1 to an 2 in S. ( S.) similis but in S. ( S.) spinosus setae ad1 and ad2 are longer than setae an1 and an2. In the material for comparison performed in this study, i.e. from the S. ( S.) similis population from Madeira and S. ( S.) spinosus from Poland, these differences were not confirmed; the shapes of sensilla and anoadanal setae are similar in both species and also setae ad1 and ad2 are the longest.
In view of a considerable morphological similarity of these two species, an attempt was made to establish their genetic relationship.
Molecular analysis of the species status of S. ( S.) crassisetosus , S. ( S.) similis , S. ( S.) spinosus , and S. ( S.) applicatus
The COI alignment for the distance calculations comprised 620 bp of unambiguous sequence data for 22 specimens of four Steganacarus species: S. ( S.) applicatus (5), S. ( S.) crassisetosus (6), S. ( S.) similis (5), and S. ( S.) spinosus (6). The number of variable sites was 207 and the estimated base frequencies were as follows: T = 0.4419, C = 0.1750, A = 0.2196, G = 0.1635. The average transition to transversion ratio (R) = 1.29 for all variable sites. NJ analysis of the COI sequences revealed four well-supported clades corresponding to the tested species (not shown). Estimates of average evolutionary divergence (K2P) were about one percent within clades grouping: S. ( S.) applicatus (1.02%, SE 0.28), S. ( S.) spinosus (1.21%, SE 0.27), and S. ( S.) similis (1.26%, SE 0.34). Nucleotide substitutions observed in the individual clades were synonymous and did not change the amino acid sequence. The highest divergence was found in S. ( S.) crassisetosus clade (3.78%, SE 0.55) but even in this case all nucleotide substitutions were synonymous. The average genetic distance among reconstructed clades was 23.03% (SE 2.15) and ranged from 20.14% (SE 1.94) between S. ( S.) crassisetosus and S. ( S.) applicatus to 27.17% (SE 2.47) between S. ( S.) spinosus and S. ( S.) similis . This level of genetic distance outnumbered intraspecific divergence and strongly support the species status of the analysed taxa. The COI nucleotide sequences were published in GenBank as DNA barcodes for S. ( S.) similis (Acc. JX891540 View Materials -43), S. ( S.) applicatus (Acc. JX891544 View Materials -48), S. ( S.) spinosus (Acc. JX891549 View Materials -54), and S. ( S.) crassisetosus (Acc. JX891555 View Materials -60).
The analysis of the nuclear D2 marker corroborated the COI results. More than five specimens from each species have been sequenced in D2 region of 28S rDNA and no intraspecific polymorphism was found among sequences from individuals belonging to the same species. The final D 2 28S rDNA alignment for the distance calculations comprised 373 nucleotide sites in which 34 were variable. The mean base frequencies were: A = 0.2347, C = 0.2043, G = 0.2737, T = 0.2872, and transition to transversion ratio (R) was 2.35. Estimates of average genetic distance over sequence pairs representing each species showed that S. ( S.) spinosus is the most diverged from the three other species (8.37%, SE 1.50%), while the average divergence among S. ( S.) crassisetosus , S. ( S.) similis , and S. ( S.) applicatus was 2.03% (SE 0.73). The genetic distances between S. ( S.) crassisetosus and S. ( S.) applicatus as well as between S. ( S.) spinosus and S. ( S.) similis did not prove unambiguously their relationship. To find out the evolutionary relations among them, a phylogenetic analysis was made on the basis of greater representation of the species belonging to Steganacaridae .
Phylogenetic relationships among S. ( S.) crassisetosus , S. ( S.) similis , S. ( S.) spinosus , S. ( S.) applicatus , and other Steganacaridae
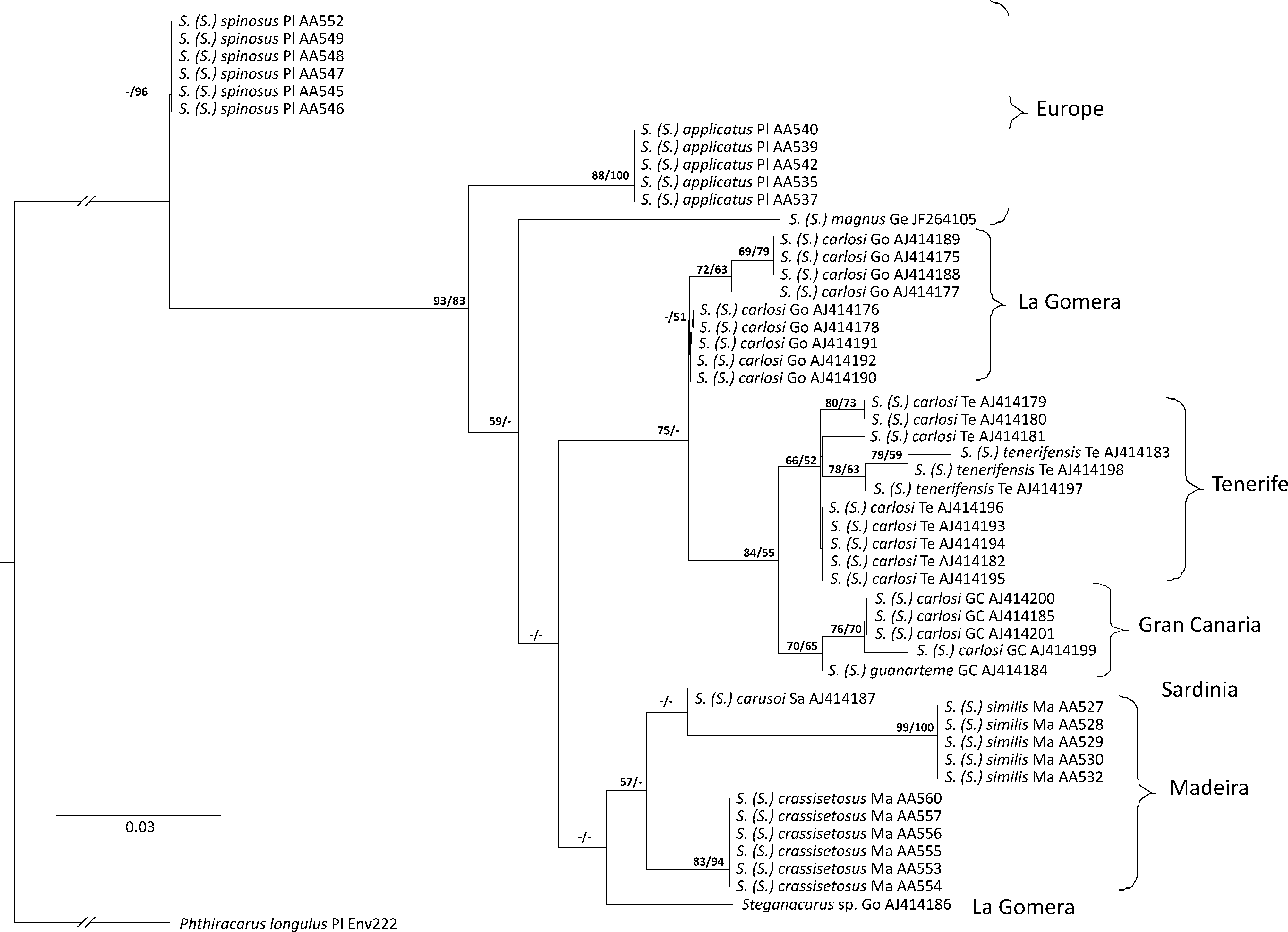
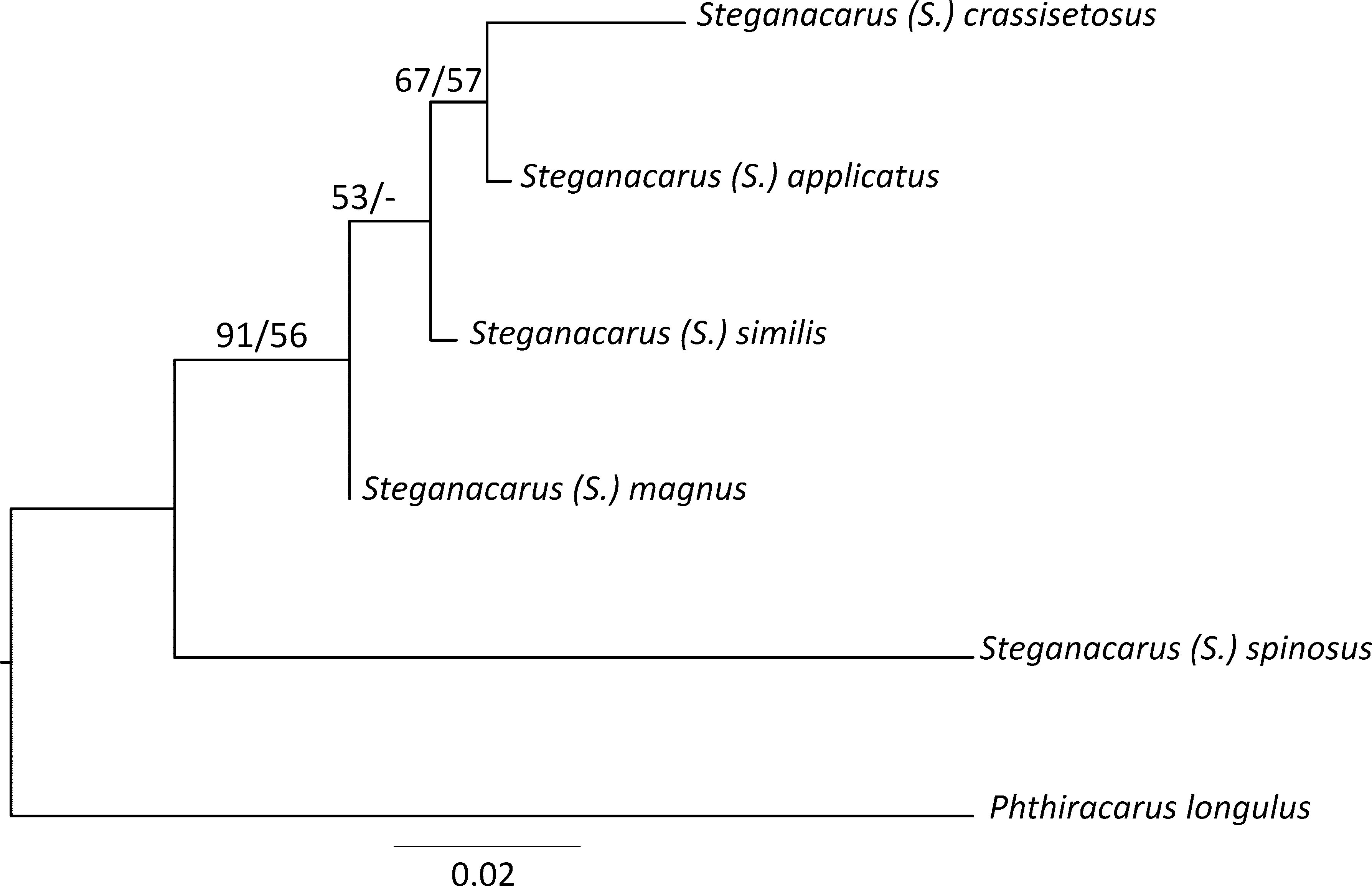
The alignment for phylogenetic tree reconstruction using COI sequence data comprised of 129 amino acid positions for 51 sequences. Twenty two sequences from this study were compared with the other known COI sequences for Steganacaridae : several species inhabiting Canary Islands and Sardinia ( Salomone et al. 2002), one representative of European species S. ( S.) magnus ( JF264105 View Materials ), and Phthiracarus longulus used as the outgroup (for details see Table 1 View TABLE 1 ). Maximum likelihood (ML) analysis based on COI amino acid data confidently recovered all analysed species ( Fig. 4 View FIGURE 4 ). Although most of the recovered evolutionary relationships among analysed Steganacaridae remain unresolved due to a low or no nodal support, the ML analysis of the COI sequences showed that S. ( S.) spinosus is the most distant species from the others and that there is no support for a close relationship between S. ( S.) applicatus and S. ( S.) crassisetosus . The ML COI tree revealed S. ( S.) spinosus basally to wellsupported (LR-ELW edge support =93, BS=83) clade containing the remainder of the taxa. In this group, European Steganacaridae were placed basally to island clades, however this placement was not supported by the tests. Canary Island species formed well-supported monophyletic clades corresponding to each island, with the exception of the position of Steganacarus sp. from La Gomera (GenBank Acc. no AJ414186 View Materials ) which grouped with two species from Madeira and one from Sardinia, but there was lack of support for this relationship. The COI amino acid sequences provided weak support (57/-) for close relationship of S. ( S.) crassisetosus and S. ( S.) similis . Both species were found in a clade with the S. ( S.) carusoi, but the latter relationship remains unresolved due to the lack of support for S. ( S.) carusoi + S. ( S.) similis clade.
The alignment for phylogenetic analysis using the sequence data from the D2 region of 28S rDNA comprised of 375 nucleotide positions for 6 species: S. ( S.) spinosus , S. ( S.) crassisetosus , S. ( S.) similis , S. ( S.) applicatus , S. ( S.) magnus , and Phthiracarus longulus used as the outgroup. The number of variable sites was 62. Phylogenetic analysis of the D2 marker performed using both the ML and NJ methods confirmed a distant relationship of S. ( S.) spinosus to the other analysed Steganacaride species. In general, the two methods generated trees with the same topology ( Fig. 5 View FIGURE 5 ) where S. ( S.) spinosus was placed basally to well-supported (BS = NJ 91%, ML 56%) clade ( S. ( S.) magnus , ( S. ( S.) similis , ( S. ( S.) crassisetosus , S. ( S.) applicatus )). Moreover, both analyses recovered with moderate support (NJ 67%, ML 65%) a closer relationship between S. ( S.) applicatus and S. ( S.) crassisetosus .
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
SubOrder |
Oribatida |
|
Family |
|
|
Genus |