ANTIPATHIDAE EHRENBERG, 1834
|
publication ID |
https://doi.org/ 10.1111/zoj.12060 |
|
persistent identifier |
https://treatment.plazi.org/id/DE793A5A-FFB2-ED4A-13AD-F90381E7FDFD |
|
treatment provided by |
Marcus |
|
scientific name |
ANTIPATHIDAE EHRENBERG, 1834 |
| status |
|
ANTIPATHIDAE EHRENBERG, 1834 View in CoL
The family Antipathidae (six genera, 116 species) is characterized by limited morphological variation in the growth form of the corallum. Colonies can be unbranched, branched irregularly, or flabellate to varying degrees, but are never pinnulate. The spines range from being simple, smooth-surfaced, triangular, and laterally compressed to conical and papillose, with a bifurcate or multiply knobbed apex (papillae: small, flattened and round to elongate striations). The polyps can vary in shape from being distinctly compressed in an axial direction to slightly elongated and are up to 3 mm in transverse diameter. The polyps of most taxa in the family have long tentacles narrowing distally to a fine tip. In fully expanded polyps, the sagittal tentacles appear longer than the lateral tentacles. Historically considered a taxonomic dumping ground, the family Antipathidae currently comprises 116 species, perhaps only a fraction of which are valid, and is thus the most species-rich family. However, the family Antipathidae is not as morphologically diverse as other families, nor does it have as great a bathymetric distribution as, for example, the Schizopathidae .
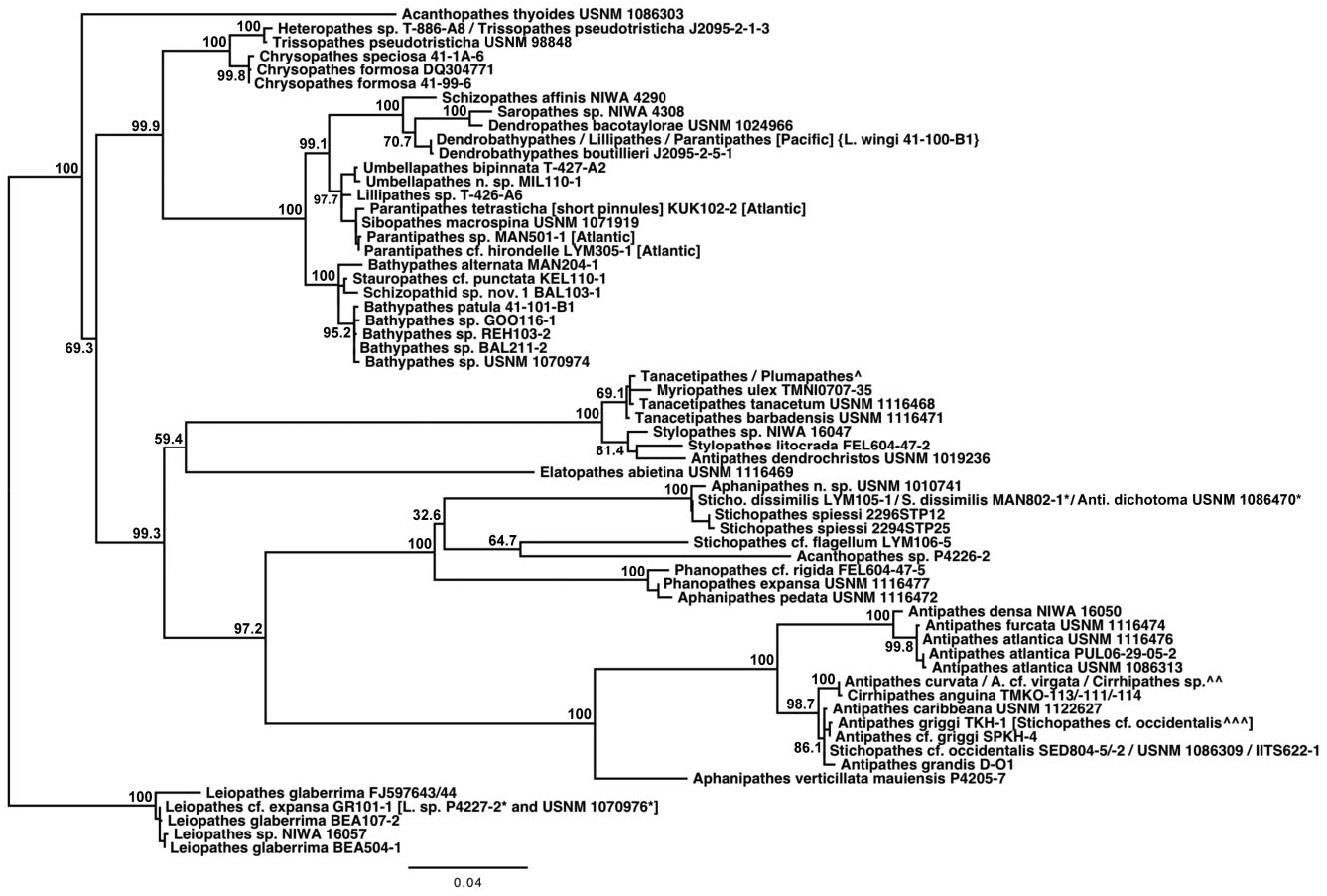
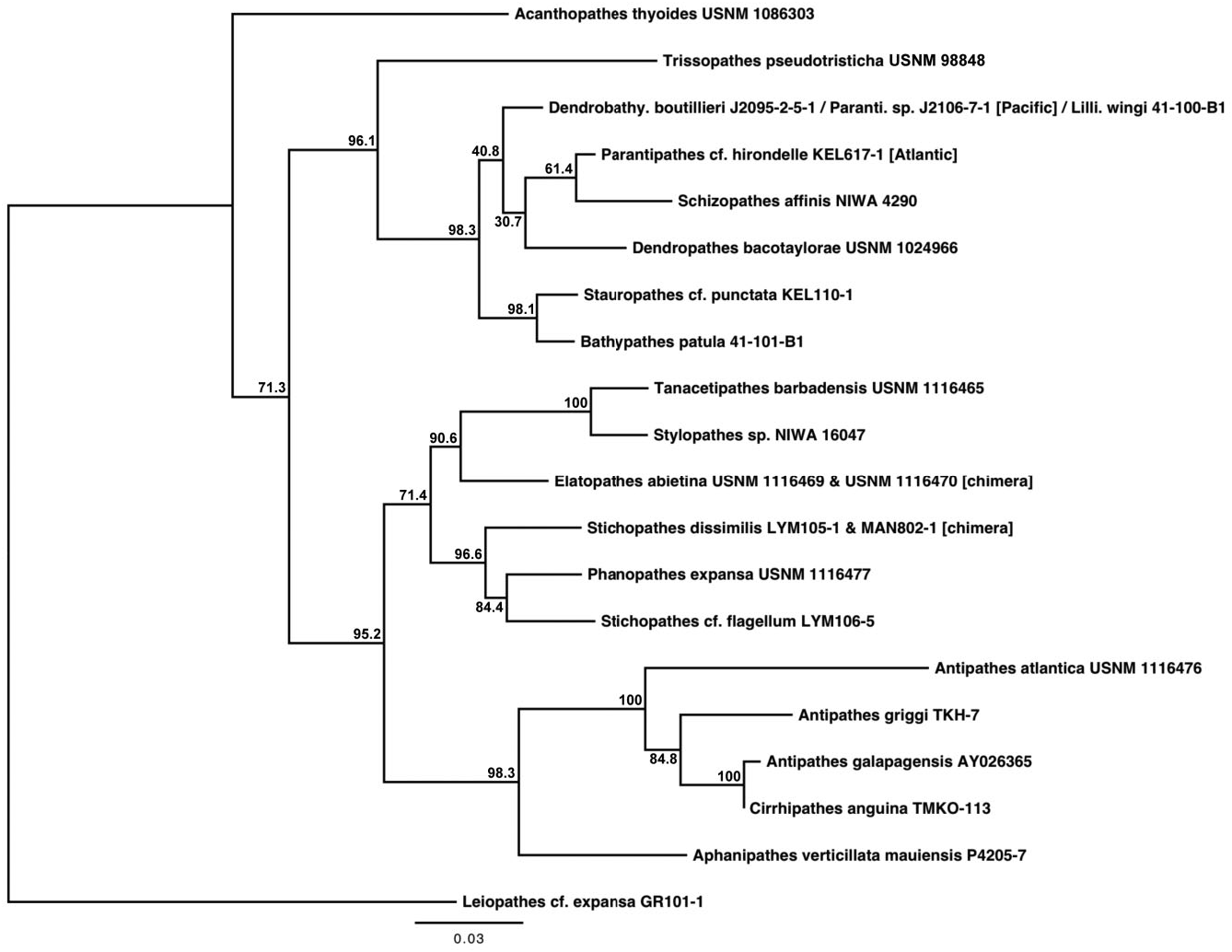
Based on morphological analyses, a minimum of 17 species, representing three genera, were obtained for sequencing, of which 12 could be referred with certainty to valid nominal species ( Table 1 & Supporting information Table S1). Based on both the mt-contig and the nuc-contig datasets, the Antipathidae was polyphyletic in all phylogenetic reconstructions. In the mt-contig phylogenies ( Figs 2 View Figure 2 , 3 View Figure 3 ), Antipathes dendrochristos grouped with the Stylopathidae while Antipathes dichotoma (the type species of the genus Antipathes ), Stichopathes dissimilis , S. cf. flagellum Roule, 1905 , and S. spiessi Opresko & Genin, 1990 all grouped with the Aphanipathidae (with the exception of A. dichotoma , these groupings are supported by morphological data; see below). Taxon sampling was more limited for the nuc-contig phylogeny ( Fig. 4 View Figure 4 ) and did not include A. dendrochristos , A. dichotoma , or S. spiessi , but S. dissimilis and S. cf. flagellum again showed a closer relationship to an aphanipathid ( Phanopathes expansa ) than to other antipathids. All subsequent references in this paper to the Antipathidae exclude these five taxa.
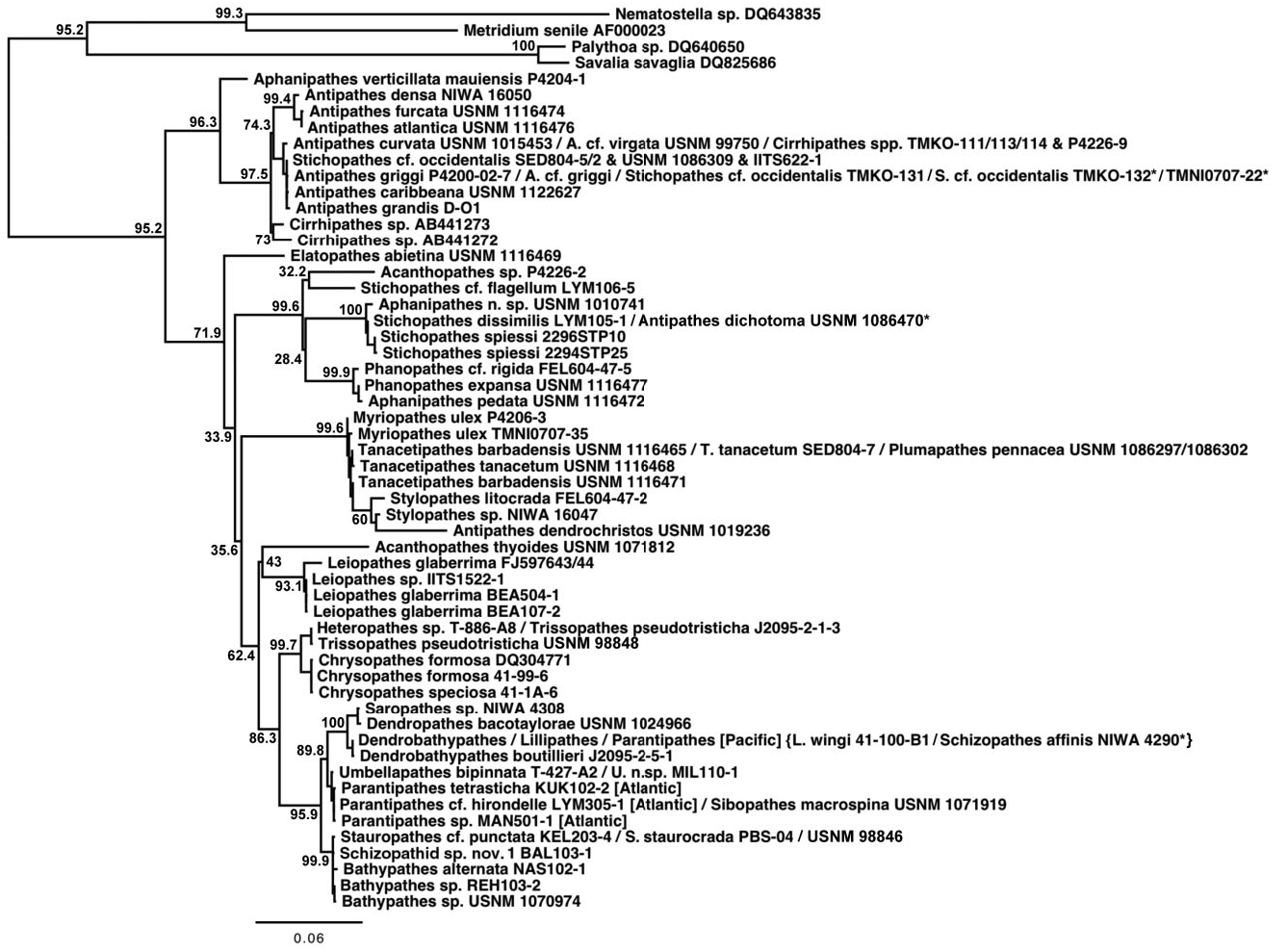
In the mt-contig phylogenies ( Figs 2 View Figure 2 , 3 View Figure 3 ), the Antipathidae is divided into two fully supported subclades, with a long branch separating Antipathes atlantica , A. densa Silberfeld, 1909 , and A. furcata from the remaining seven species (five Antipathes and one each of Stichopathes and Cirrhipathes ). Morphologically, these subclades are distinguished by the corallum (somewhat flabellate or fan-shaped in the former and unbranched or irregularly branched in the latter) and different spine morphologies (smooth, compressed triangular spines in the former, and more conical, often knobby and/or papillose spines in the latter). Two sequences downloaded from GenBank and identified as Cirrhipathes sp. (collected in Kenting, Taiwan) formed a third subclade in the cox 3- cox 1 phylogeny ( Fig. 5 View Figure 5 ), distinct from the Hawaiian Cirrhipathes obtained for this study (specimens identified by D.M.O.). However, one of our Cirrhipathes colonies (P4-226-9) shared the same multilocus mt haplotype as specimens of A. curvata van Pesch, 1914 and A. cf. virgata Esper, 1788 (but no sequence was obtained for igrW for A. cf. virgata ). All of these genetically identical colonies were collected in the tropical/sub-tropical North Pacific, but span a distance from Hong Kong to the Galapagos. A diagnostic character for the genus Cirrhipathes is that colonies are unbranched. However, the polyphyly of the genus and their relationship to branching Antipathes species suggests that branching pattern might not be a valid synapomorphy, as has been noted among calcaxonian octocorals ( France, 2007). On the other hand, in both Hawaiian Cirrhipathes sp. and A. curvata , there is a tendency for the polyps to occur in multiple rows. Polyp organization as a diagnostic character may be worth further analysis among genera in the Antipathidae .
A further illustration of the problem with using ‘non-branching’ as a genus-level diagnostic character is seen for Stichopathes . Like Cirrhipathes , Stichopathes is also polyphyletic: Stichopathes cf. occidentalis grouped within the Antipathidae but S. dissimilis , S. cf. flagellum , and S. spiessi cluster with the Aphanipathidae (these will be discussed further in the Aphanipathidae section). Both groups of Stichopathes include specimens collected from the Atlantic and Pacific, but those in the Antipathidae are from relatively shallow water (<100 m depth), whereas those clustering with the Aphanipathidae are strictly deep sea (490–1681 m depth). Also like Cirrhipathes , the different species of Stichopathes are more closely related to branching Antipathes or aphanipathids than to each other or the unbranched Cirrhipathes .
Grouping sister to the Antipathidae in all six phylogenetic reconstructions was Aphanipathes verticillata mauiensis . Given that the close sister relationship of Aphanipathes verticillata mauiensis to the bulk of the Antipathidae was strongly supported in all mtDNA and nuclear DNA phylogenies, two genetic distance estimates were determined for the family Antipathidae : estimates including and excluding Aphanipathes verticillata mauiensis . Additional taxon sampling is necessary to determine if Aphanipathes verticillata mauiensis should be classified in the Antipathidae or Aphanipathidae . Although quite divergent from one another, the Antipathidae (along with Aphanipathes verticillata mauiensis ) grouped sister to a clade of Aphanipathidae with strong support (BS: 97.2–98.5; BPP: 99–100).
Although taxon sampling was limited, the results of the nuc-contig phylogenies were largely congruent with the results of the mt-contig phylogenies: the Antipathidae formed a polyphyletic assemblage, with S. dissimilis and S. cf. flagellum grouping with the aphanipathid Phanopathes expansa , and Antipathes spp. and Cirrhipathes grouping sister to Aphanipathes verticillata mauiensis (BS: 98.3; BPP: 100). However, unlike the mtDNA results, the Antipathidae , along with Aphanipathes verticillata mauiensis , grouped sister to a clade comprising three families (BS: 95.2; BPP: 100); this three-family sister clade placed the Aphanipathidae sister to the Stylopathiade + Myriopathidae (BS: 71.4; BPP: 72–99). Due to incomplete taxon sampling within the Antipathidae in the nuclear DNA dataset, there was no clear distinction between the two subclades of Antipathidae that were revealed by the mtDNA dataset; however, the branch leading to A. atlantica was considerably longer than branches leading to other antipathids.
Whereas all phylogenetic reconstructions (except cox 3- cox 1 and 18S) were rooted with the Leiopathidae , the cox 3- cox 1 and 18S phylogenies (rooted with sea anemones and zoanthids) suggested that future phylogenetic reconstructions should also consider using the Antipathidae as the root; however, care should be taken when choosing antipathids as the family has historically been a polyphyletic dumping ground. The cox 3- cox 1 nucleotide-based phylogenetic reconstruction ( Fig. 5 View Figure 5 ), which also revealed a polyphyletic Antipathidae , grouped the Antipathidae (not including Antipathes dendrochristos , which grouped with the Stylopathidae , or A. dichotoma and some Stichopathes spp. that grouped with the Aphanipathidae ) sister to the remaining antipatharian families. With the exception of Elatopathes abietina , the Antipathidae clade was the least divergent ingroup relative to the outgroups. The cox 3- cox 1 amino acid-based phylogenetic reconstruction (not shown) and 18S phylogenies (not shown) revealed similar results. As noted above, an unpublished phylogeny by the CnidToL Working Group of the entire phylum Cnidaria based on concatenated mt rnl, 18S and 28S sequence data placed Leiopathes fully supported sister to all other antipatharians. Thus, there was a conflict in phylogenetic signal among the various gene regions analysed. Moving forward, genes that are found in a true outgroup (e.g. anemones or zoanthids) must be explored to determine whether the Antipathidae or Leiopathidae is the true sister to the remaining antipatharians.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |