Limosilactobacillus reuteri, 2021
|
publication ID |
https://doi.org/10.1099/ijsem.0.004644 |
|
DOI |
https://doi.org/10.5281/zenodo.6310193 |
|
persistent identifier |
https://treatment.plazi.org/id/CD6F3526-FFC9-252B-443B-FC00FE32248B |
|
treatment provided by |
Felipe |
|
scientific name |
Limosilactobacillus reuteri |
| status |
subsp. nov. |
DESCRIPTION OF LIMOSILACTOBACILLUS REUTERI SUBSP. RODENTIUM SUBSP. NOV.
Limosilactobacillus reuteri subsp. rodentium (ro.den′ ti.um. L. pl. gen. n. rodentium of gnawing animals, reflecting adaptation of the subspecies to rodents).
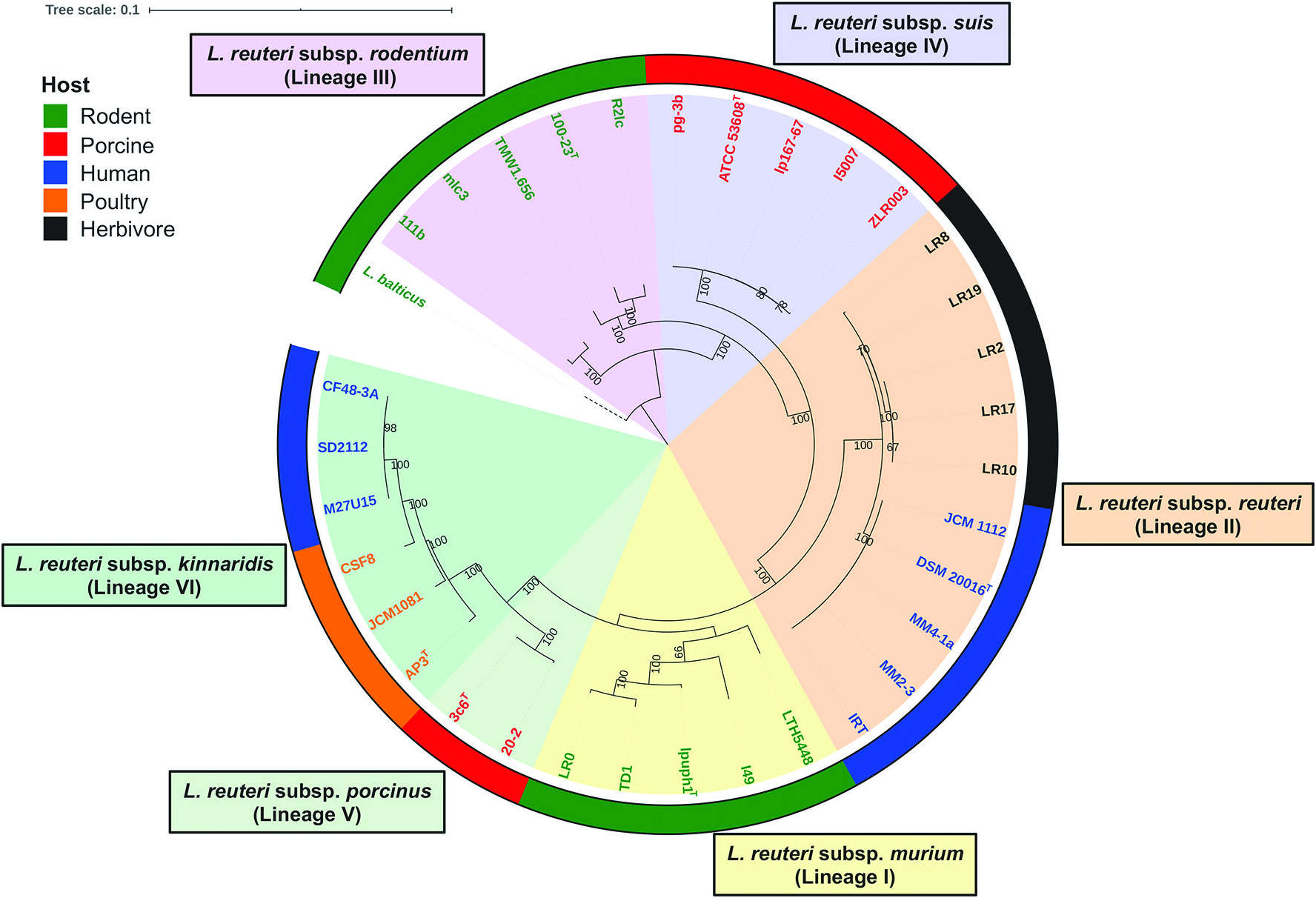
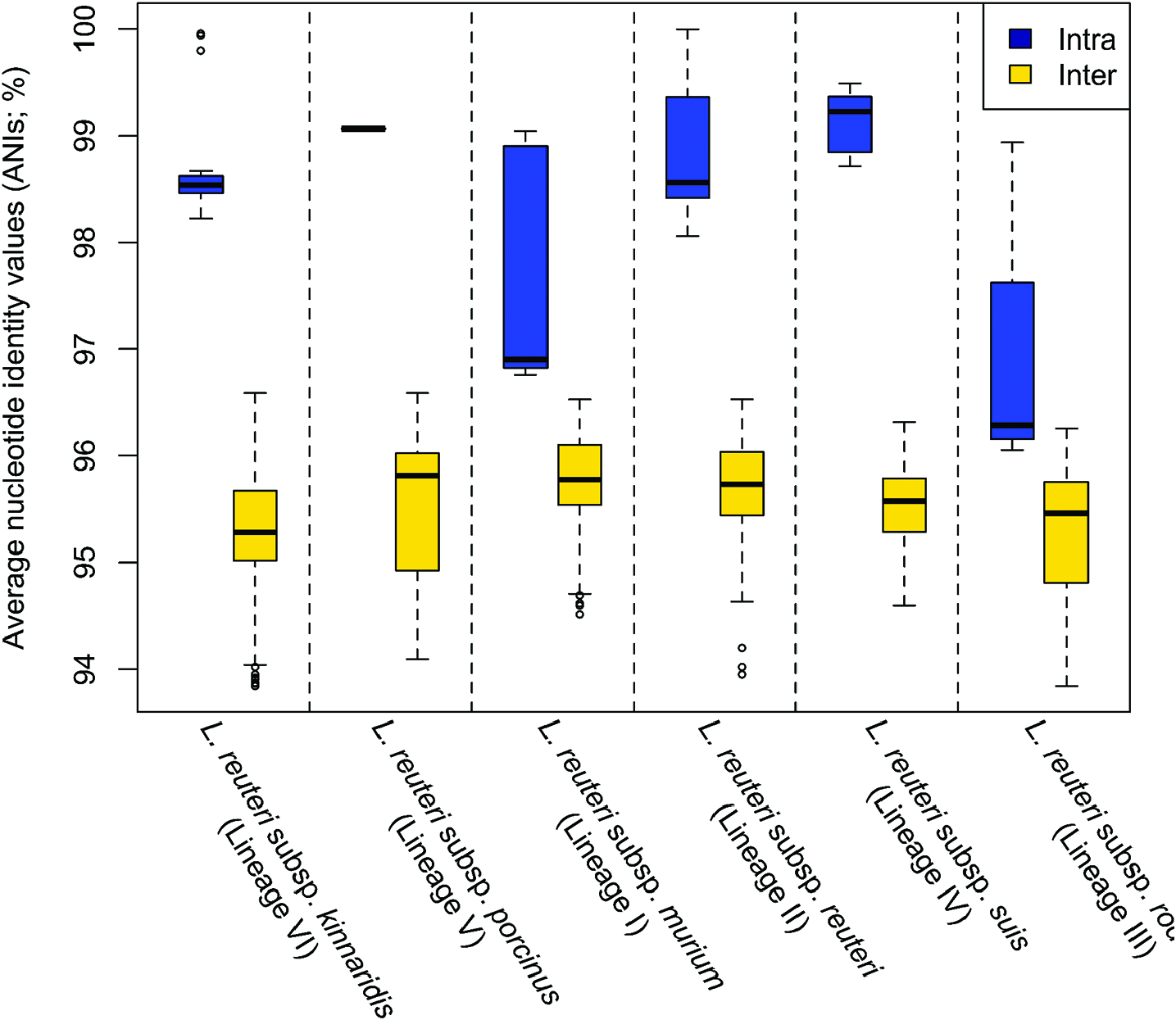
L. reuteri strains clustered in lineage III ( Fig. 3 View Fig ) belong to L. reuteri subsp. rodentium and were mainly isolated from rodents [ 5 – 7]. Strains of this subspecies have ANI values of 96.1–98.9 % with each other and ANI values of 93.8–96.3% with other L. reuteri strains belonging to different subspecies ( Fig. 4 View Fig ). Acid is produced from D-ribose,D-galactose, D-glucose, maltose, lactose, melibiose, sucrose, raffinose and potassium gluconate; acid production from L-arabinose and D-xylose is strain-specific; acid is not produced from D-fructose, D-mannose, methyl α- D-glucopyranoside, aesculin, glycerol, erythritol, D-arabinose, L-xylose, D-adonitol, methyl β -D-xylopyranoside, L-sorbose, L-rhamnose, dulcitol, inositol, D-mannitol, D-sorbitol, methyl α- D-mannopyranoside, N -acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, trehalose, inulin, melezitose, starch, glycogen, xylitol, gentiobiose, turanose, D-lyxose,D-tagatose,D-fucose,L-fucose,D-arabitol, L-arabitol, potassium 2-ketogluconate or potassium 5-ketogluconate. Phylogenetic analyses based on the core genes identified in this study ( Fig. 3 View Fig ) and a previous study [ 5], AFLP and MLSA (using concatenated sequences of ddl, pkt, leuS, gyrB, dltA, rpoA and recA genes) [ 7] indicate that strains clustered in this lineage including the sourdough isolates are rodent-specific. Strains of L. reuteri subsp. rodentium displayed elevated fitness in mice through the colonization and biofilm formation on the forestomach epithelium [ 5, 7, 11], suggesting adaptive evolution with rodents that led to host specificity. Large surface proteins (>750 aa) exist among strains belonging to this subspecies, which involve in epithelial adhesion and biofilm formation [ 6]. A xylose operon is highly conserved for this subspecies, especially for strains originating from rodents [ 6], and thus most strains of this subspecies could metabolize xylose that is an important substrate for gut bacteria [ 51]. In addition, strains of this subspecies produce the enzyme urease for acid resistance and rarely produce the antimicrobial compound reuterin [ 6, 8]. Sourdough isolates of this subspecies (LTH2584, TMW1.106, TMW1.112 and TMW1.656) produce reutericyclin, a unique antimicrobial tetramic acid with activity against Gram-positive bacteria [ 52].
The type strain, 100-23 T (=DSM 17509 T =CIP 109821 T), was isolated from the rat gastrointestinal tract [ 11, 53], with a DNA G+C content of 38.7mol%.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
Limosilactobacillus reuteri
| Li, Fuyong, Cheng, Christopher C., Zheng, Jinshui, Liu, Junhong, Quevedo, Rodrigo Margain, Li, Junjie, Roos, Stefan, Gänzle, Michael G. & Walter, Jens 2021 |
L. reuteri
| SUBSP. RODENTIUM 2021 |
L. reuteri subsp. rodentium
| Li & Cheng & Zheng & Liu & Quevedo & Li & Roos & Gänzle & Walter 2021 |
L. reuteri
| SUBSP. RODENTIUM 2021 |
L. reuteri subsp. rodentium
| Li & Cheng & Zheng & Liu & Quevedo & Li & Roos & Gänzle & Walter 2021 |