Gracilipsodes similis, WARD (2001)
|
publication ID |
https://doi.org/ 10.1111/j.1096-3642.2008.00403.x |
|
persistent identifier |
https://treatment.plazi.org/id/C046B71F-CB38-0D28-FCA5-5D18F9FF8F32 |
|
treatment provided by |
Felipe |
|
scientific name |
Gracilipsodes similis |
| status |
|
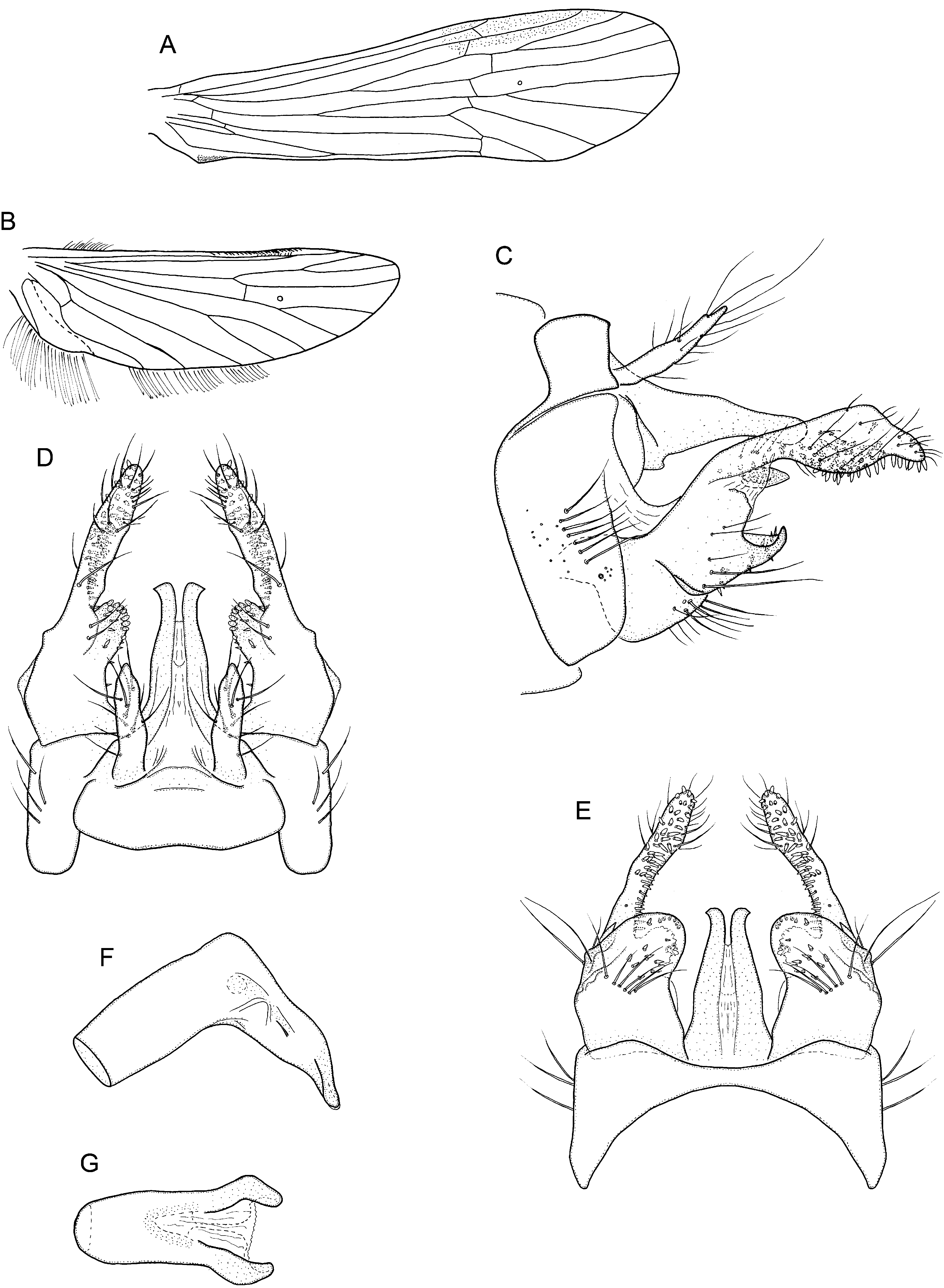
GRACILIPSODES SIMILIS WARD (2001) ( FIGS 10 View Figure 10 , 13 View Figure 13 )
Gracilipsodes similis Ward (2001: 76–81) .
Diagnosis: The species is distinguished from all other species in the genus, except G. psocopterus , by having the tibial spur formula 0, 1, 1, and by a darker body and forewing colour. It differs from G. psocopterus by the shorter superior appendages, by the shallower split between the tergum X processes, and by the shape of the apical part of the inferior appendage.
Description, male: Body medium brown to dark greyish brown. Forewing brown to greyish brown. Tibial spur formula 0, 1, 1. Hind tibiae each with 12–13 small spines (N = 2). Forewing ( Fig. 10A View Figure 10 ): length 5.5–6.1 mm (N = 3); apex broadly rounded; forks 1 and 5 present; crossvein m–cu about as long as or slightly shorter than crossvein r–m, oriented at a sharp angle to M. Hindwing ( Fig. 10B View Figure 10 ): length 4.1– 4.4 mm (N = 2); apex rounded; forks 1 and 5 present; crossvein r–m straight; costa with 25–30 hamuli.
Genitalia: Generally strongly sclerotized, dark. Segment IX annular, laterally setose; in lateral view, sternite IX nearly rectangular, broad, tergite IX half as wide as sternite IX ( Fig. 10C View Figure 10 ); tergite IX posterior margin weakly produced posteroventrad in dorsal view ( Fig. 10D View Figure 10 ), in ventral view, sternum IX median margin widely and shallowly ellipsoid ( Fig. 10E View Figure 10 ). Superior appendages setose, flattened dorsoventrally, shorter than tergum X, apices nearly acute in dorsal view ( Fig. 10D View Figure 10 ). Tergum X strongly sclerotized, divided into two lateral spines connected with semipermeable membrane to distal two-thirds; in lateral view, basal part wide, tapering to rounded apex, with small ventral process basally, ( Fig. 10C View Figure 10 ); in dorsal and ventral views, apices curving laterally ( Fig. 10D, E View Figure 10 ); pre-apical lateral processes absent. Inferior appendages each bipartite, setose; basal part broad in ventral and lateral views ( Fig. 10C, E View Figure 10 ), mesally concave, produced posteromesally into a broad, apically rounded lip, bearing small dorsal spines ( Fig. 10C, E View Figure 10 ); apical part of each inferior appendage digitate, almost straight in dorsal view ( Fig. 10D View Figure 10 ); in lateral view, widening slightly at mid length, apex slender, with 40–50 mesoventral spines ( Fig. 10C View Figure 10 ); mesal process distinct, with 10–12 strong, rounded mesal spines ( Fig. 10D View Figure 10 ). Harpago: small, strongly sclerotized, ovate ( Fig. 10C View Figure 10 ). Phallic apparatus simple ( Fig. 10F, G View Figure 10 ); phallicata basally tubular, fused with phallobase, apex narrow; sharply bent ventrally at mid length, apex with a pair of sclerotized, lateral processes, being produced posteroventrad, curved somewhat medially, with apex narrowly rounded ( Fig. 10F,G View Figure 10 ); phallotremal sclerite U-shaped ( Fig. 10G View Figure 10 ).
Material examined: One male, New Caledonia, Province Nord, Amoa River, 23 m a.s.l., 25–26 November 2001, light trap, loc#20 (Johanson, Pape & Viklund); 13 males, New Caledonia, Province Nord, 50 m upstream of bridge on Hienghène-Tnèdo road, 3.9 km south of summit of Mt Tnèda , 2.4 km east of Tnèdo , 20°43.099′S, 164°50.108′E, 27 m a.s.l., 7 December 2003 – 1 January 2004, loc#069 (Johanson) GoogleMaps .
Distribution: Eastern parts of Province Nord.
PHYLOGENETIC RESULTS
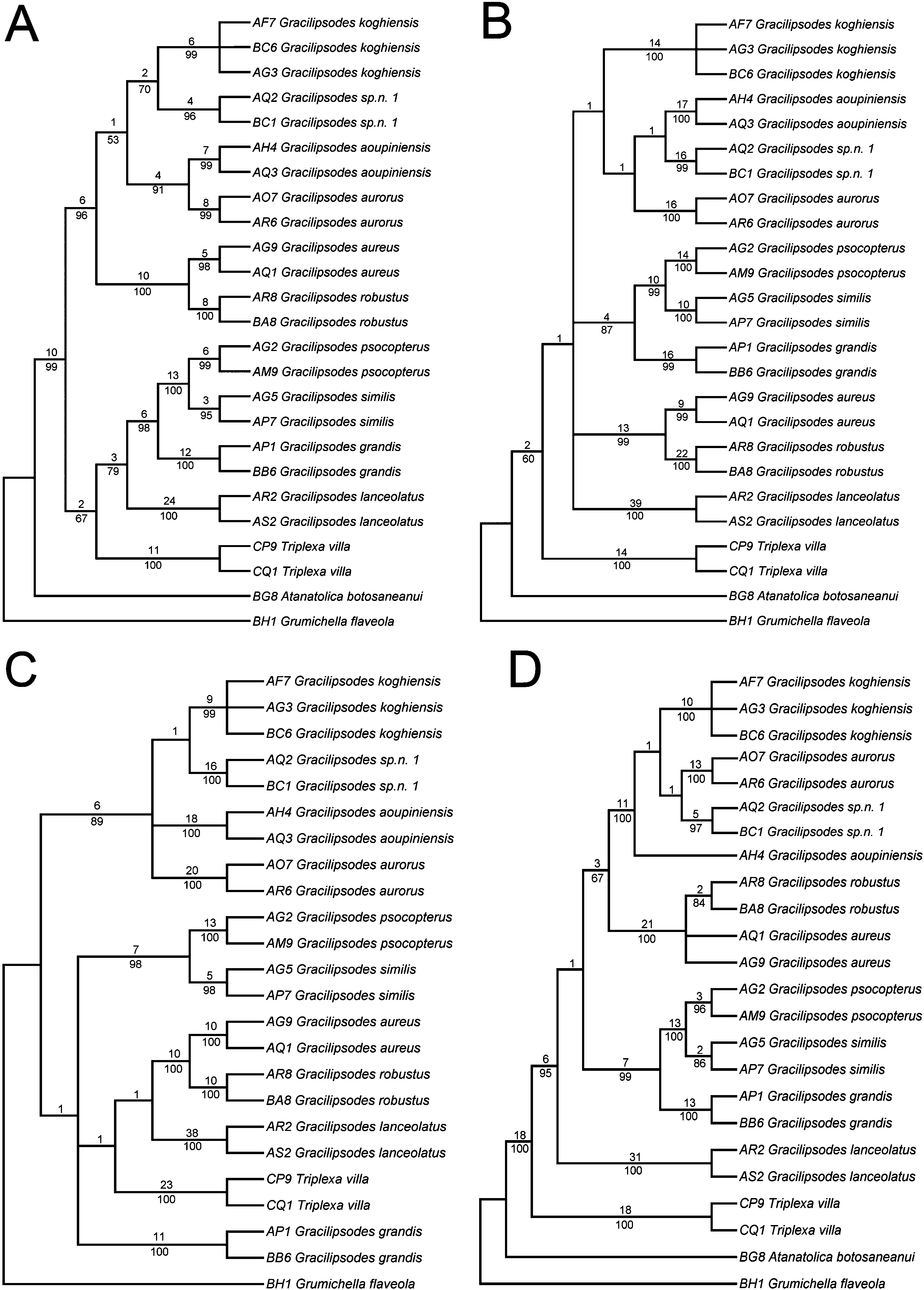
In all analyses, the individual Gracilipsodes species formed monophyletic units with jackknife support of 84–100% (except for G. aureus in the EF-1a analysis, which collapses basally of G. robustus ), but the relationship among the various species differed between the genes analyzed. The gene producing the overall lowest branch support was EF-1a.
The implied alignment of the mitochondrial ribosomal 16S gene fragments yielded 380 homologous characters, of which 87 were parsimony informative for all 25 specimens. P distances are shown in Table 3, indicating the high similarity between G. similis and G. psocopterus , as well as for G. koghiensis and G. sp. nov. 1. The subsequent parsimony analysis of this gene alone resulted in one shortest tree (L = 252) ( Fig. 11A View Figure 11 ). We recovered Triplexa basally in the Gracilipsodes species clade consisting of ( G. lanceolatus , ( G. grandis , ( G. psocopterus , G. similis ))). This relationship has, however, low branch support (jackknife = 67%). This clade again forms the sister clade to the second major clade containing the three species pairs ((( G. robustus , G. aureus ), (( G. aurorus , G. aoupiniensis ), (G. sp. nov. 1, G. koghiensis ))).
The mitochondrial COI data set consisted of 658 characters, of which 212 were parsimony informative for all 25 specimens. According to the P distances in Table 4, the highest similarity is found between G. psocopterus and G. similis . The parsimony analysis of this data set yielded two shortest trees (L = 640) ( Fig. 11B View Figure 11 ), placing Triplexa as sister to all Gracilipsodes . This position was poorly supported (jackknife = 60%). The basal divergence within Gracilipsodes is poorly resolved, but holds two strongly supported clades ( G. aureus , G. robustus ) and ( G. psocopterus , G. similis ), both with jackknife support of 99%.
The data set of the mitochondrial COII gene fragment was one taxon shorter than for the other genes, resulting from the absence of a sequence from A. botosaneanui , and included 512 characters, of which 176 were parsimony informative. According to the P distances in Table 5, the highest similarity is found between G. similis and G. psocopterus , as well as for G. aureus and G. robustus . The parsimony analysis of this character set resulted in three shortest trees (L = 494) ( Fig. 11C View Figure 11 ), with conflicts between them assigned basally within Gracilipsodes , resulting in poor resolution at this level. Here again we found Triplexa inside Gracilipsodes as sister to [ G. lanceolatus , ( G. aureus , G. robustus )], but this configuration had a low jackknife support (below 50%). As in the results of the COI dataset analysis, the results here strongly support the two clades ( G. aureus , G. robustus ) and ( G. psocopterus , G. similis ) as being monophyletic.
The nuclear EF-1a data set excluded one specimen because of an absent sequence (the AQ3 G. aoupiniensis ), and consisted of 1099 characters, of which 194 were parsimony informative for all 24 individuals. According to Table 6, the P distances show very high sequence similarity within the species pairs G.
Species set in bold are sp. nov.
‘n.a.’ indicates only one specimen present within the species, thus no distance span.
Species set in bold are sp. nov.
‘n.a.’ indicates only one specimen present within the species, thus no distance span.
Species set in bold are sp. nov.
‘n.a.’ indicates missing sequence or only one specimen present within the species, thus no distance span.
Species set in bold are sp. nov.
‘n.a.’ indicate only one specimen present within the species, thus no distance span.
aureus + G. robustus and G. psocopterus + G. similis . The parsimony analysis yielded one shortest tree (L = 440) ( Fig. 11D View Figure 11 ), where Triplexa was found to form the sister group to Gracilipsodes . The monophyly of Gracilipsodes received a high jackknife value (95%). The individuals of G. lanceolatus were grouped together and formed the sister group to all the remaining Gracilipsodes species. The remaining species formed two major clades, one with low jackknife support (67%) – ((( G. aureus , G. robustus ), ( G. aoupiniensis , ( G. koghiensis , ( G. aurorus , G. sp. nov. 1)))) – and one with strong jackknife support (99%) – ( G. grandis , ( G. psocopterus , G. similis ). Here, all species are moderately to strongly supported, except G. aureus , which collapses basally of G. robustus .
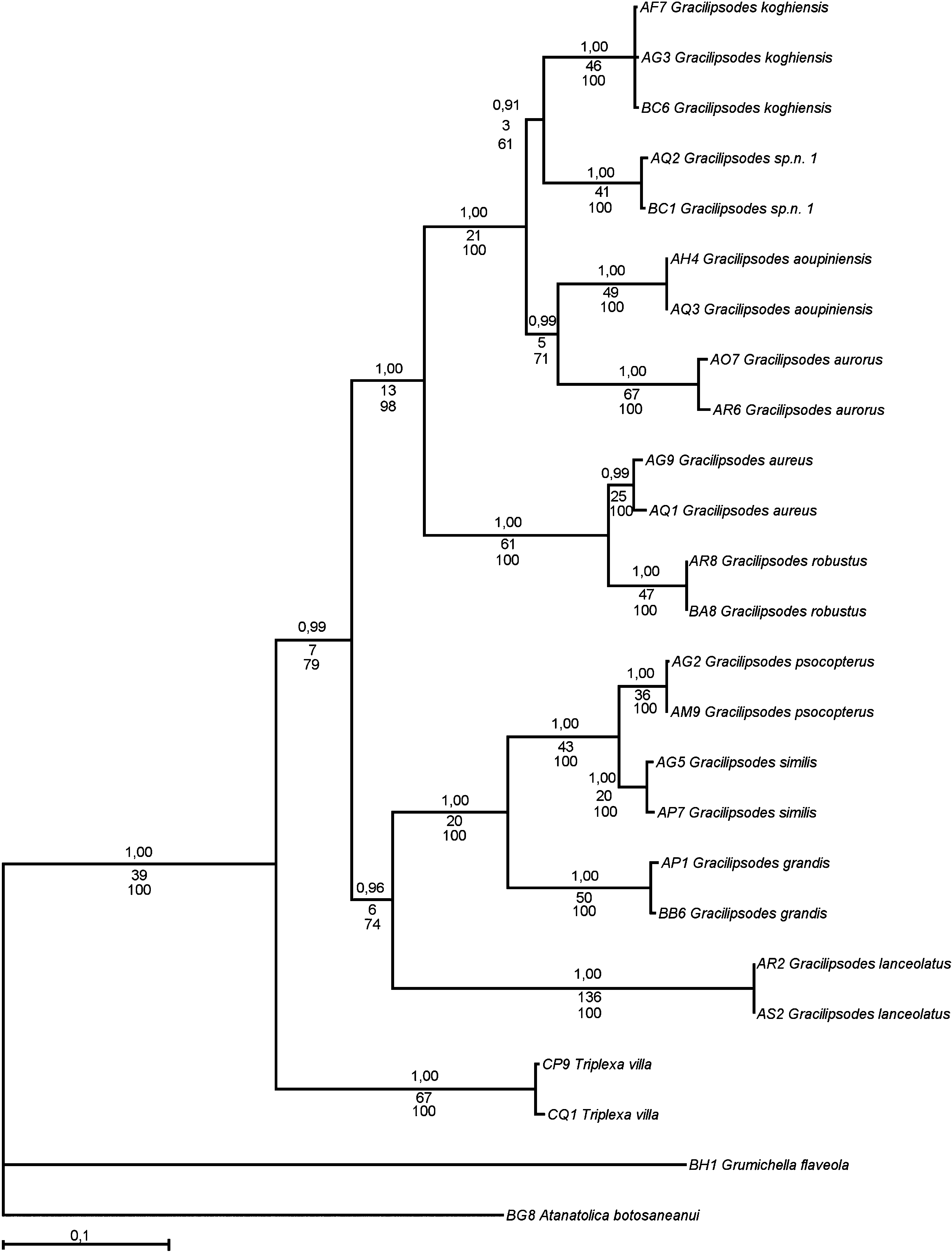
The combined data set yielded a matrix consisting of 25 individuals and 2649 characters (of which 670 were parsimony informative). The parsimony analysis resulted in a single shortest tree (L = 1839) ( Fig. 12 View Figure 12 ), where Triplexa formed the sister group to Gracilipsodes (as in the results of the EF-1a and COI analyses). The monophyly of Gracilipsodes received a jackknife support of 79%, and the genus was divided into two major clades: ( G. lanceolatus , ( G. grandis , ( G. psocopterus , G. similis ))) and (( G. aureus , G. robustus ), (( G. koghiensis , G. sp. n. 1), ( G. aoupiniensis , G. aurorus ))). Most of the nodes received strong support, except 61% for (( G. koghiensis , G. sp. nov. 1), ( G. aoupiniensis , G. aurorus )), 71% for ( G. aoupiniensis , G. aurorus ), and 74% for ( G. lanceolatus , ( G. grandis , ( G. psocopterus , G. similis ).
The data-set analysis in MrModeltest ( Nylander, 2004) for the separate genes gave no single model for all gene partitions, but the two genes COI and COII shared the ‘GTR + invgamma’ model, EF-1a was proposed to be analyzed with ‘GTR + gamma’, and 16S was proposed to be analyzed with ‘HKY + invgamma’. These models were assigned to their respective dataset partition, and were then run in MrBayes in a combined analysis. This analysis resulted in a tree topologically identical to the most parsimonious tree generated from the combined analysis ( Fig. 12 View Figure 12 ). All nodes received high posterior probability values (± 0.99), except the G. koghiensis + G. sp. nov. 1 clade and the G. lanceolatus – G. similis clade, with posterior probabilities of 0.91 and 0.96, respectively.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |
Gracilipsodes similis
| Malm, Tobias & Johanson, Kjell A. 2008 |
Gracilipsodes similis
| Ward JB 2001: ) |