Jemasonia pater ortizi (Orellana, [2010]) Zhang, Jing, Cong, Qian, Shen, Jinhui, Song, Leina, Orellana, Andrés, Brockmann, Ernst, Mielke, Carlos G. C., Mielke, Olaf H. H., Costa, Mauro & Grishin, Nick V., 2023
|
publication ID |
https://doi.org/ 10.11646/zootaxa.5319.4.7 |
|
publication LSID |
lsid:zoobank.org:pub:AE993A70-D5C2-46E3-A570-40EF0DB7ECC2 |
|
DOI |
https://doi.org/10.5281/zenodo.8211470 |
|
persistent identifier |
https://treatment.plazi.org/id/AF07C417-FFAD-2D58-15F7-FB2A90EAFF07 |
|
treatment provided by |
Plazi |
|
scientific name |
Jemasonia pater ortizi (Orellana, [2010]) |
| status |
stat. nov. |
Jemasonia pater ortizi (Orellana, [2010]) , stat. nov.
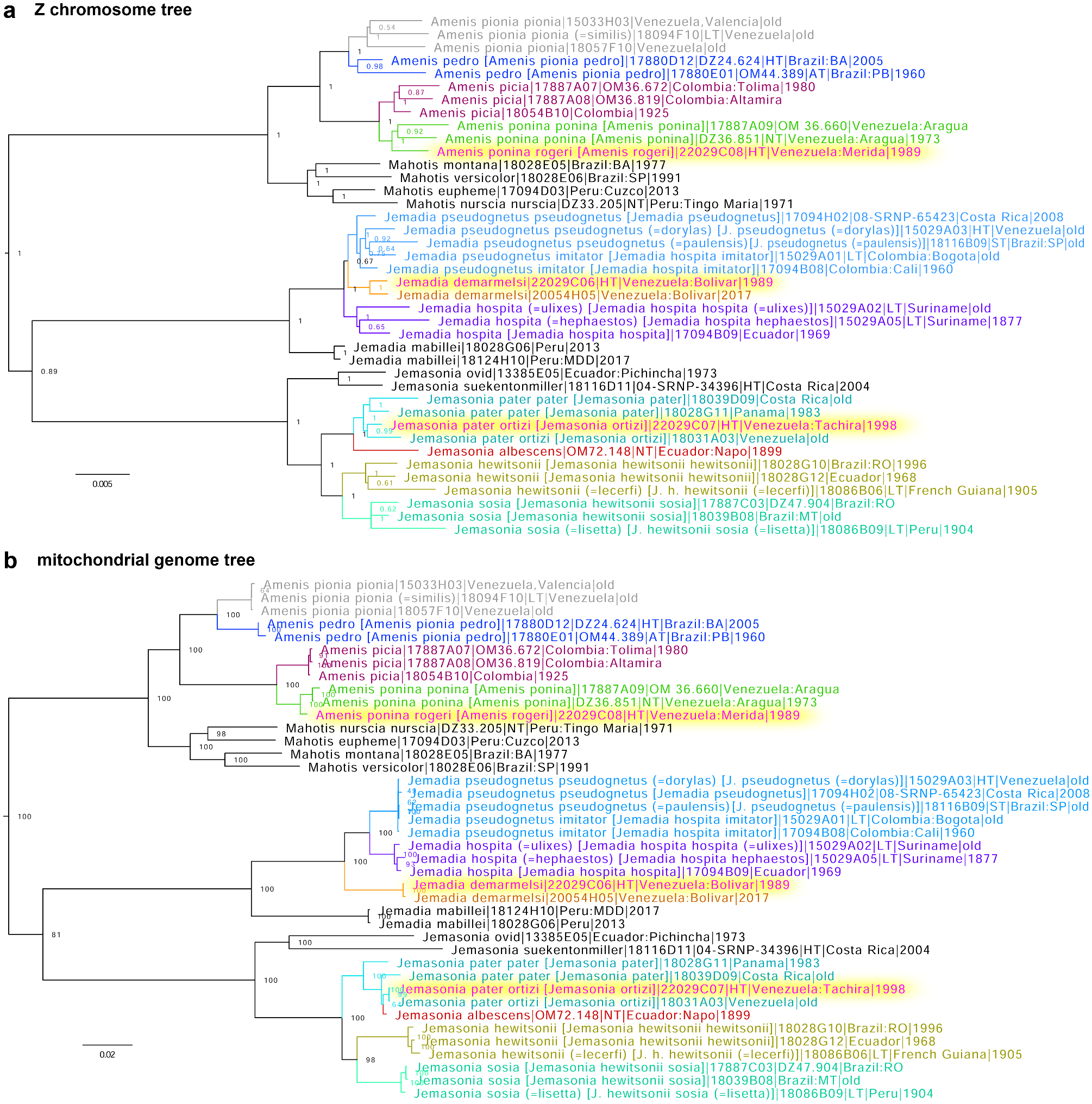
Genomic analysis reveals close clustering of the holotype of Jemadia ortizi Orellana, [2010] (type locality in Venezuela: Táchira) in the MIZA collection with specimens of Jemasonia pater ( Evans, 1951) (type locality in Colombia: Cundinamarca) ( Fig. 1 View FIGURE 1 ). Their COI barcodes could be 100% identical in some specimens. As stated (and illustrated) in the original description, genitalia of J. ortizi are the same as J. pater (“Igual a los de J. pater ”) ( Orellana [2010]), with the major difference between the two taxa being in the dark palpi and darker body coloration beneath. Taxa differing in coloration but similar in genomes and genitalia are traditionally regarded as subspecies. Therefore, we propose to treat J. ortizi as a subspecies of J. pater : Jemasonia pater ortizi (Orellana, [2010]) , stat. nov. The COI barcode sequence of the holotype of J. p. ortizi , sample NVG-22029C07, GenBank accession OR178495, 658 base pairs is:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAATTGGAACATCCCTTAGATTACTAATTCGAACTGAATTAGGAACTCCAGAATCTTTAATTGGA GATGATCAAATTTATAATACTATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGAGGATTTGGAAATTGACTAG TTCCTCTAATATTAGGAGCACCTGATATAGCTTTCCCCCGAATAAATAACATAAGATTTTGATTATTACCCCCTTCATTAACTTTACTTATTTCAAGAAG CATTGTAGAAAATGGTGCCGGAACTGGATGAACAGTTTACCCCCCTCTTTCCTCTAATATCGCCCACCAAGGAGCTTCTGTAGATTTAGCTATTTTTTCT TTACATTTAGCTGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACAATTATCAATATACGAATTAAAAACTTATCTTTTGATCAAATACCTT TATTTGTTTGAGCCGTAGGAATTACAGCATTATTATTACTTTTATCATTACCTGTATTAGCAGGAGCTATTACTATATTATTAACAGATCGAAATATTAA TACTTCTTTTTTTGATCCTGCTGGAGGAGGTGATCCTATTTTATATCAACATTTATTT
This sequence is identical to the barcode of Jemasonia albescens (Röber, 1925) neotype (DZ72.148), suggesting a possibility of introgression between these species, because in the Z chromosome tree, the neotype is sister to J. pater being separated from it by a branch with the length characteristic of closely related but distinct species ( Fig. 1a View FIGURE 1 ). Their genitalia also differ ( Grishin et al. 2014; Mielke et al. 2022).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |