Lathyrus sativus, L.
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2022.113296 |
|
DOI |
https://doi.org/10.5281/zenodo.8262727 |
|
persistent identifier |
https://treatment.plazi.org/id/8825E574-2775-5368-A16A-BD80FC378063 |
|
treatment provided by |
Felipe |
|
scientific name |
Lathyrus sativus |
| status |
|
2.1. Lathyrus sativus suspension culture system
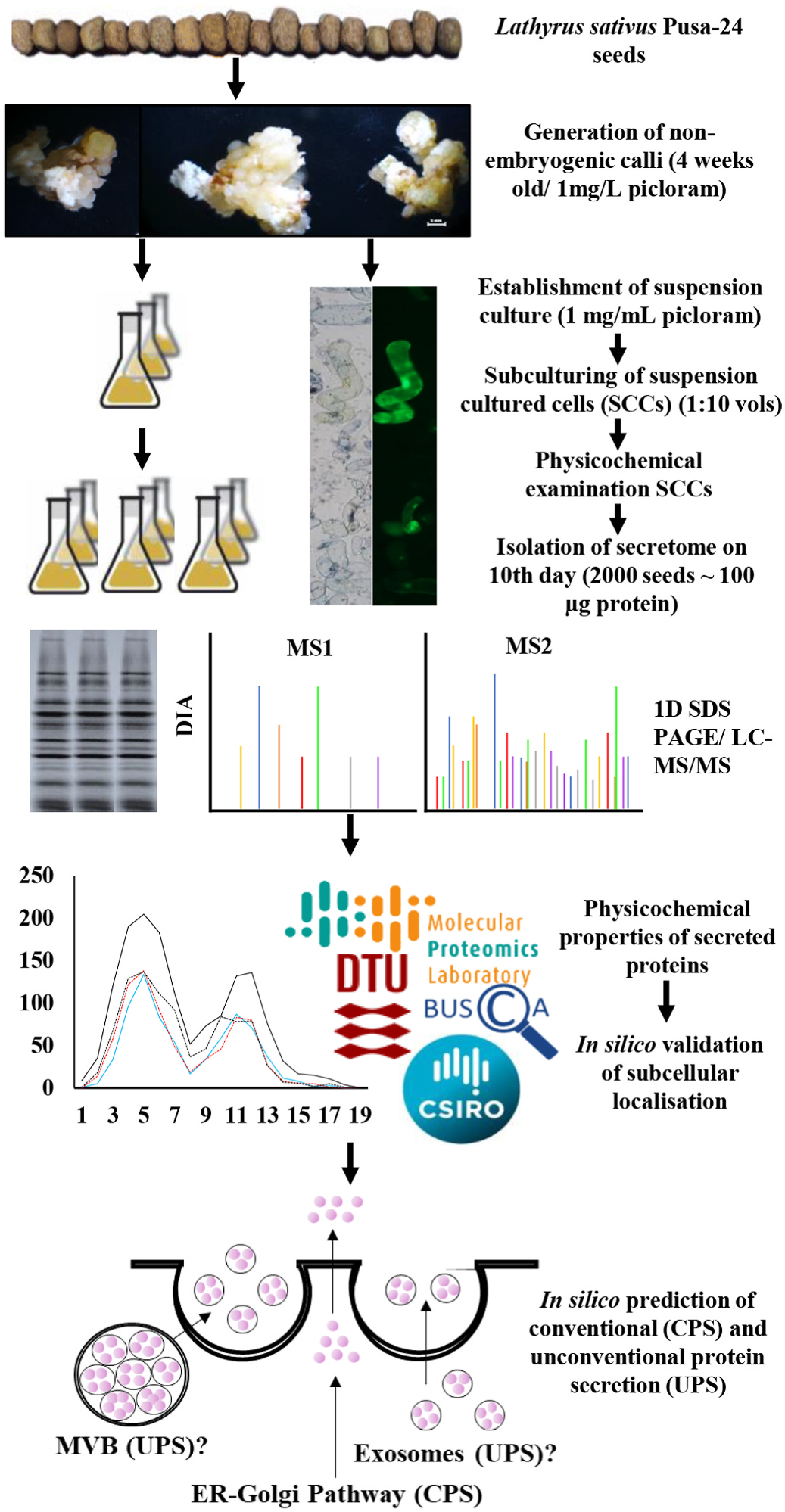
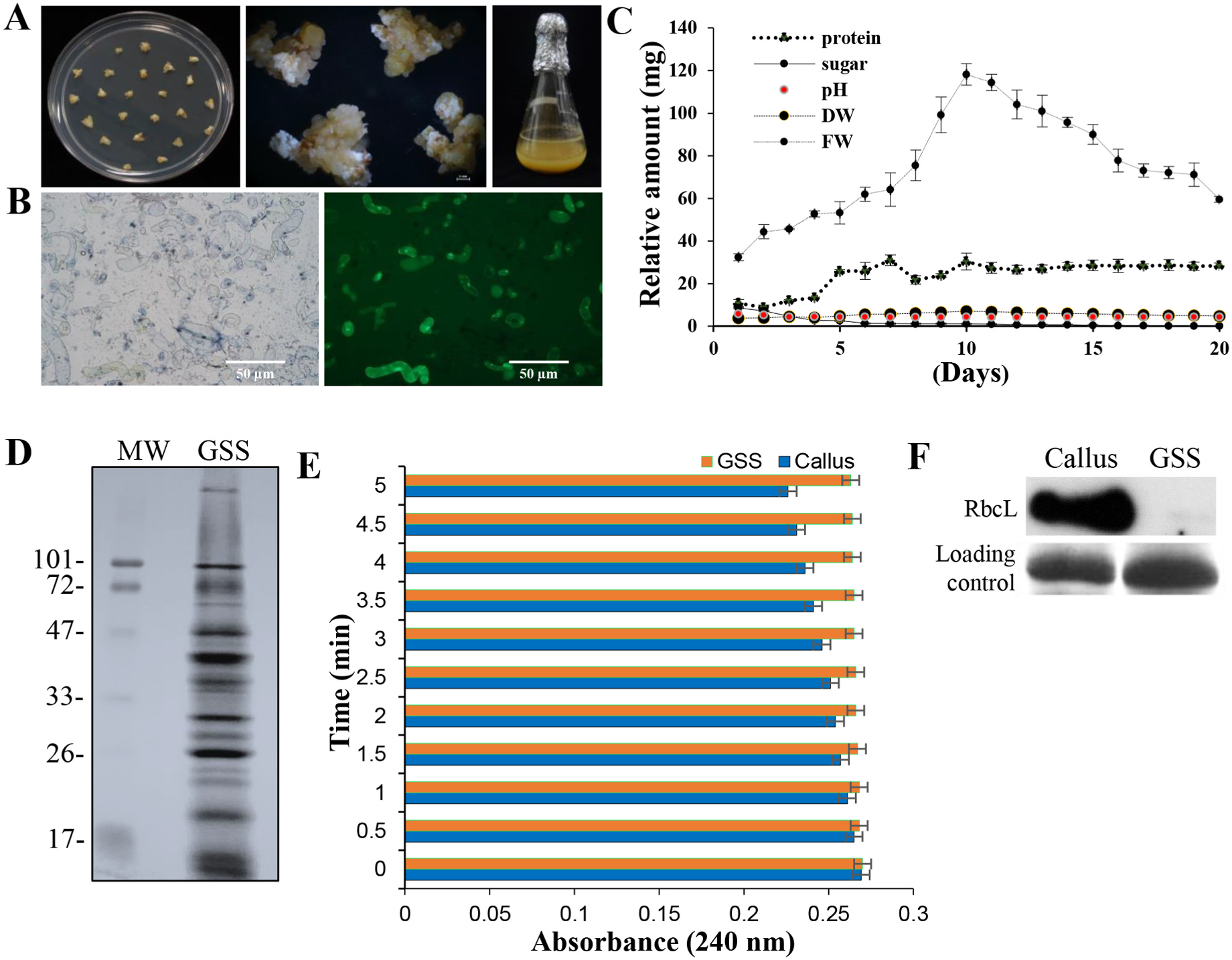
Dedifferentiated plant SCCs constitute a totipotent single cell-type homogenous system, which is more effective for investigating biological functions and regulatory mechanisms in the context of developmental stimuli than tissue-enriched multicellular systems. Among crops, legumes are an interesting set of plants that express unique secretory proteins, particularly in nodulation-induced root samples. Hence, we prepared the non-embryogenic calli of an orphan legume, L. sativus , for generating totipotent cells and developed the reference secretome map. The overall research design and flow process are shown in Fig. 1 View Fig . The resulted SCCs served as a defined set of grasspea secreted proteins, which were employed for the identification of species-specific “secretome markers”. Friable, non-embryogenic and cream colored calli were obtained using 1 mg /mL picloram ( Fig. 2A View Fig ) over other phytohormone combinations, which induced organogenesis ( Rathi et al., 2019a). The calli so obtained ranged from 20 mg in 2nd week up to 100 mg in 4th week of growth. Four-week-old calli were used to successfully establish SC. Live cells stained green with fluorescein diacetate (FDA), while the dead cells stained blue with Evans blue ( Fig. 2B View Fig ). The SCCs were of different shapes, such as small round cells, large round cells, ellipsoids and elongated cells, conforming to the different growth stages of totipotent cells. Physiological assessments were performed after the second subculturing to determine the suitable stage for the application of stress ( Fig. 2C View Fig ). While the total protein content was found to increase, the soluble sugars decreased in the SCCs, reflecting active growth ( Fig. 2C View Fig ). The biomass and viability of SCCs were increased until the 10th day, beyond which these parameters decreased ( Fig. 2C View Fig ). The pH of suspension cultures stabilized at 4.2 after the 3rd day of subculturing, reflecting the active growth of SCCs ( Fig. 2C View Fig ). Therefore, the 10th day of culture was an ideal stage for harvesting the secretome.
2.2. Purity and enrichment of the secretome
The secretome samples from 3 biological replicates were pooled, concentrated, and visualized by CBB staining ( Fig. 2D View Fig ). The GSS gel profile displayed a mixed proportion of moderate and low molecular weight proteins. Furthermore, the purity assessment of the GSS fraction revealed minimal contamination with cytosolic components ( Fig. 2E and F View Fig ; Supplementary Fig. S1 View Fig ). The catalase activity was observed to be multifold higher in the total callus protein sample compared to GSS (p-value <0.05) ( Fig. 2E View Fig ). Immunoblot analysis using the chloroplastspecific RubisCo large subunit (RbCL) revealed a negligible presence of rbcL in GSS compared to callus samples ( Fig. 2F View Fig ). Overall, GSS was essentially free from cytosolic and chloroplast contamination.
2.3. Identification and predicted localization of secreted proteins
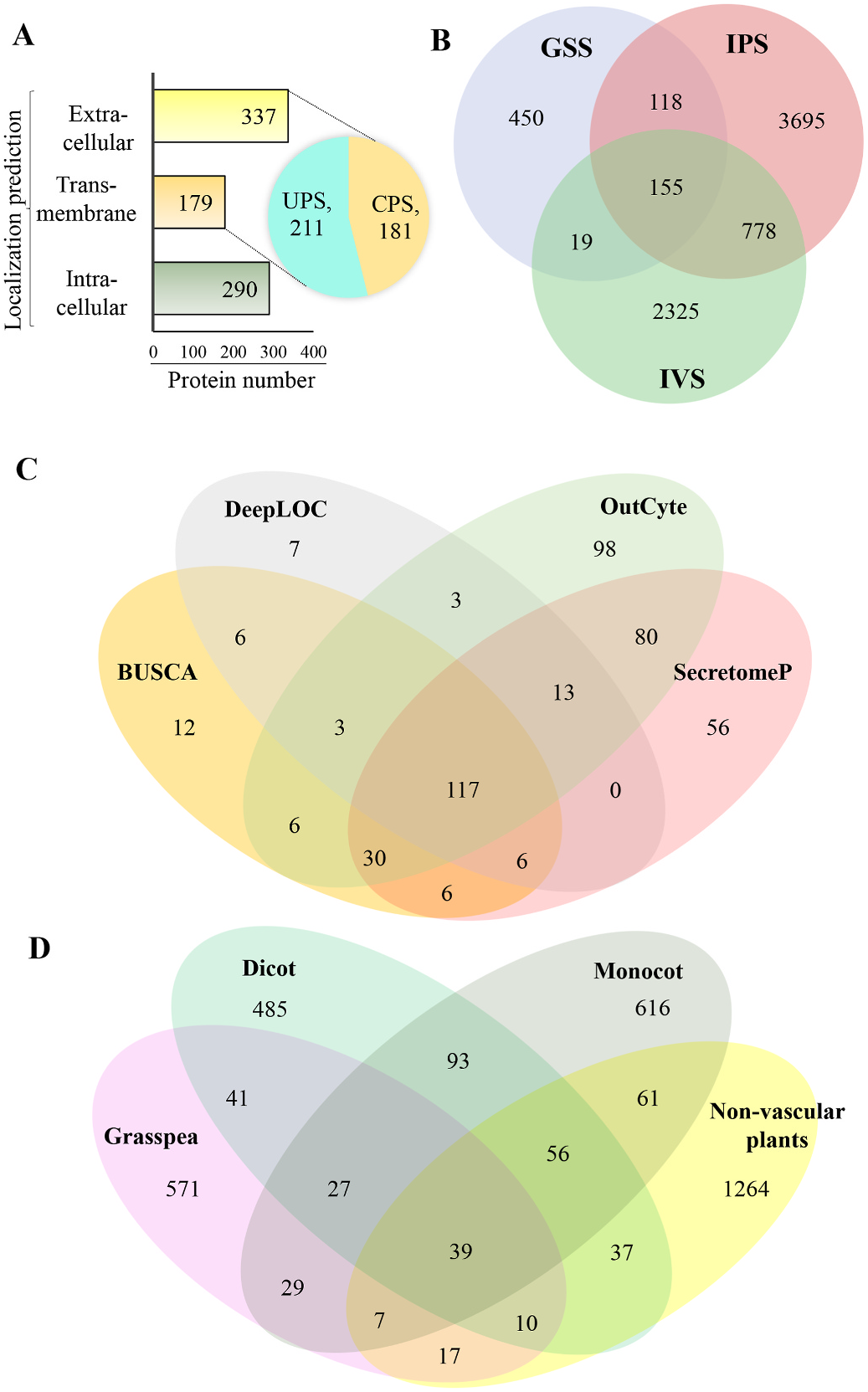
The LC-MS/MS analysis of GSS yielded a nonredundant set of 741 proteins ( Fig. 3A and B View Fig ; Supplementary Table S1). Thirty-four proteins associated with plasmodesmata, physical mediators of cell–cell signaling in plants, were identified in our study, including the dual specificity protein kinase YAK1, germin-like proteins, nectarin-1, subtilisin-like protease SBT2.1, peroxidase 71, LRR receptor-like serine/ threonine-protein kinase At 1g 14390, SUMO-activating enzyme subunit 2 and pinoresinol reductase 2, among others. Eighty-two cell wallassociated proteins were identified, including cellulose synthase-like protein D5 and catalytic subunits 11 and 4, reduced wall acetylation 4 and NAC domain-containing protein 10, among others. Although protein secretion is a tightly regulated process, we observed the secretion of intracellular proteins as reported previously ( Jeffery, 2016). One of the ENTH/VNT family proteins, clathrin interactor Epsin 2, which is known for intercellular membrane trafficking, was also found to be secreted into the grasspea extracellular space. Other examples include nucleotide binding proteins such as pumilio (APUM-5), FRS12 (FAR1-RELATED SEQUENCE12), DNA-directed RNA polymerase and ATP-dependent RNA helicases.
The present repertoire of the grasspea secretome highlights the functional divergence of the plant secretome. The secreted proteins mediate signaling not only in actively growing regions of the plant body ( Hu et al., 2021) but also in processes related to plant reproduction ( Chae and Lord, 2011; Matthys-Rochon, 2005). Significantly, a myriad of functional protein classes was identified in our study that have direct roles in conferring biotic ( Haegeman et al., 2012) and abiotic stress responses ( Nakaminami et al., 2012). Disease resistance proteins (S145, S211, S372, S412, S455, S468, S672, S704) were characteristically expressed in the GSS samples and were previously identified to be effective against fungi, viruses and nematodes ( McDowell et al., 1998; Cooley et al., 2000; Zhang and Gassmann, 2007; Yuan et al., 2018; Warmerdam et al., 2020). The disease resistance-like protein dominant suppressor of CAMTA3 (DSC1) plays a significant role in conferring basal immunity in Arabidopsis ( Warmerdam et al., 2020) . The secretion of arginyl transferase 1 (ATE1) is particularly intriguing, since posttranslational arginylation of secreted signaling proteins is of immense significance across the varied kingdoms of life ( Saha and Kashina, 2011). ATE1 was even identified as a constituent of exosome-like vesicles in Taenia asiatica ( Liang et al., 2019) . Targeting fungal virulence factors is an effective plant defense strategy, and in the present study, we identified proteins of similar functionality, including Kiwellin-1 ( Altegoer et al., 2020). BOI-related E3 ubiquitin-protein ligase, a crucial plant defense protein against biotic and abiotic stress, was also observed in the GSS. The cell defense-related UBP1-associated protein 2B (UBA2) ( Kim et al., 2008) was secreted in GSS samples, raising questions about its role in the extracellular space, since its role in the nucleus is associated with mRNA stabilization ( Lambermon et al., 2002). Genes associated with abiotic stress tolerance included the SNARE-interacting proteins KEULE and syntaxin ( Kwon et al., 2020). One of the PERK family genes, proline-rich extensin-like receptor kinase 5 (AtPERK5), was also a constituent of the grasspea suspension secretome. PERKs have been indicated to be critical for not only abiotic stress tolerance but also plant growth and development ( Borassi et al., 2016; Kesawat et al., 2022). Two late embryogenesis abundant proteins (LEA14 and LEA76) were active constituents of the GSS profile, and LEA proteins have established roles in desiccation tolerance ( Candat et al., 2014). The secretion of auxin response factor 5 (Monopteros) is also intriguing, as previous studies have established its role in regulating mobile proteins ( Schlereth et al., 2010), but none have reported Monopteros to be extracellular. One of the water-deficit responsive transcription factors, bHLH45 ( Wang et al., 2018), was also secreted in L. sativus . An abiotic stress tolerance-promoting PLAT domain-containing protein ( Hyun et al., 2014), was also identified in our study.
The secreted proteins were sequentially subjected to localization prediction analysis using 7 different programs. SignalP and SecretomeP predicted the presence of signal peptides in 147 and 183 proteins, respectively, and 38 were exclusively predicted by the latter. SecretomeP also predicted 123 proteins to be secreted through the UPS. We found an additional increase in the prediction of secreted proteins using OutCyte, wherein 139, 133 and 75 were categorized as CPS, UPS and TM, respectively. Interestingly, two proteins (S468, probable disease resistance protein and S687, cytochrome f) were predicted to follow UPS by OutCyte but CPS by SecretomeP. While ApoplastP predicted 69 proteins as bona fide ECM constituents, BUSCA and DeepLoc predicted 140 and 112 proteins as ECM residents, respectively. TM domains were predicted in 152 GSS proteins by TMHMM. Furthermore, organellardefined predictions were made by DeepLoc and BUSCA, which predicted 43 and 46 proteins to be PM residents, respectively. ApoplastP and SignalP predicted a redundant set of GSS proteins to be secreted compared to other programs. Only 117 GSS proteins were commonly predicted by BUSCA, DeepLOC, OutCyte and SecretomeP ( Fig. 3C View Fig ). Approximately 60% of the proteins were predicted to be secreted, as projected with the variable softwares. Furthermore, ~25% of proteins were predicted to follow CPS, and ~29% were predicted to follow UPS. Since OutCyte, SecretomeP and BUSCA predicted the maximum number of secreted proteins, we utilized these programs in the study of crop secretomes in a conjunctive manner.
Although there is recent evidence for the secretion of cytoplasmic constituents to the cell surface to assume different biological functions ( Zininga et al., 2018), this field of study demands more experimental evidence, even more so in the case of plants. Since the dawn of the plant secretome, plasma membrane proteins have been frequently identified in the outermost fraction, presumably owing to their functionality as cell surface-associated proteins ( Tanveer et al., 2014; Nogueira-Lopez et al., 2018; Ramulifho et al., 2019; Flores-Tornero et al., 2021; Ngcala et al., 2020; Vincent et al., 2020). Membrane proteins often contain a defined signal peptide but are secreted through the UPS, as in the case of cystic fibrosis transmembrane conductance regulator (CFTR) ( Gee et al., 2011). Experimental evidence of non-classically secreted plasma membrane proteins has yet to be ascertained in plants ( Robinson et al., 2016). Additionally, the localization of TM domain-containing transporters and ion channels is frequently determined by active processes of exocytosis and endocytosis, which is reflected in the dynamic composition of the eukaryotic secretome. Moreover, the UPS pathway attributes functional divergence to known cytoplasmic proteins, for instance, the Hsps ( Campanella et al., 2014; Reddy et al., 2018). This family of proteins functions as chaperones intracellularly but behaves more as signaling proteins extracellularly. The possible routes of UPS have not yet been completely elucidated ( Ding et al., 2014), except for dissection of the exosome pathway and compositional modulation in response to environmental constraints ( Woith et al., 2021). Also, synaptotagmin mediated UPS as well as intercellular communication has been established in plants ( Zhang et al., 2011; Uchiyama et al., 2014) A cytosolic cellular signaling protein, translationally controlled tumour protein (TCTP), was found to be secreted in L. sativus , like its homolog in tobacco pollen tubes ( Hafidh et al., 2016).
2.4. Novel and known domains of the Lathyrus sativus secretome
We conducted an in-depth analysis of the identified proteins, highlighting the presence of both known and novel domains in GSS (Supplementary Table S1; Supplementary Fig. S2 View Fig ). The 14-3-3 proteins (S33, S61) were identified in GSS, corroborating their indispensable presence in the plant secretome ( Wen et al., 2007; Gupta et al., 2011; Liu et al., 2015; Ves-Urai et al., 2021). Hydroxyacid dehydrogenases identified in GSS were previously reported to be a part of fungal ( Severino et al., 2014; Fang et al., 2021) and microbial secretomes ( Mastronunzio et al., 2009; Komeil et al., 2014). Eight AAA domain-containing proteins (S19, S171, S197, S360, S384, S542, S583, and S600) were identified in the GSS, like the secretomes of sunflower ( Pinedo et al., 2012) and model moss species ( Lehtonen et al., 2014), and these are associated with ATPase-associated cellular activities. Abhydrolase domain-containing proteins comprised 2-hydroxyisoflavanone dehydratase, auxin response 4 and methylesterase 4, and these were observed to follow CPS and UPS. These proteins were previously reported to be a part of insect and fungal secretomes ( Pandey et al., 2018; Zhu et al., 2020) and known to function in a UPS-dependent manner ( Garcia-Ceron et al., 2021). Aminotransferases ( Cheng et al., 2009; Wang et al., 2014; Williams et al., 2014) and ATP-synthase domain-containing proteins ( Agrawal et al., 2010; Espino et al., 2010; Vincent et al., 2020) have long been known to be part of global eukaryotic secretomes. Further, the cysteine-rich secretory proteins (CAP superfamily) play crucial roles in biomolecular interactions in the extracellular space ( Schneiter and Di Pietro, 2013), and four of these proteins were identified, including three PR-1s and a P14A. Copper oxidases (S305, S500, S555) are crucial components of eukaryotic secretomes, as they aid in conferring mechanical stiffness to the ECM ( Lum and Min, 2011; Ricard-Blum and Vallet, 2019). Interestingly, cupin superfamily proteins were particularly abundant (30 proteins) in the GSS, in accordance with previous plant secretome reports. Cupins are relevant in the ECM and/or secretome owing to their functions in the modification of mannose and rhamnose residues in higher plant cell walls ( Dunwell et al., 2001).
DEAD-box helicases (S198, S446, S618) were identified in GSS despite their nucleus-specific functions. Interestingly, these proteins have also previously reported in insect ( Etebari et al., 2020) and fungal secretomes ( Liu et al., 2015; Dussert et al., 2019). Dirigent domain-containing proteins (S211, S294, S391) indicated their function as abiotic ( Ngcala, 2018) and biotic stress-responsive constituents ( Regente et al., 2017; Haseeb et al., 2021) of the extracellular space. Proteins harboring domains of unknown function (DUF1604, DUF296, DUF677, DUF4378, DUF825, DUF295, DUF825, DUF1744, DUF1298, DUF588) appeared to be predominant constituents of the grasspea secretome (Supplementary Table S1). Among these, DUF296 was previously reported to be differentially regulated in the cell wall of wheat endosperm ( Mehdi et al., 2020). Hsps have now been recognized as ‘moonlighting proteins’ in the extracellular space, especially Hsp70 and Hsp90, which are mostly secreted through the UPS ( Agrawal et al., 2010; Rabouille, 2017). GDSL lipases have previously been reported in Medicago and Arabidopsis secretomes ( Oh et al., 2005; Kusumawati et al., 2008), and noticeably, lipases were also identified in the GSS. Similarly, maturase K (7 GSS proteins) has been reported in the extracellular space of pitcher plant ( Zakaria et al., 2019), sea buckthorn ( Sougrakpam and Deswal, 2016), sunflower ( Pinedo et al., 2012) and tobacco ( Millar et al., 2009). Also, p450 (11), peroxidases (17), protein kinase (24), pentatricopeptide repeat (PPR) domain-containing proteins (14), thaumatin (4), thioredoxin (4), U-box (5) and WD40 (7) were abundantly represented in the GSS, in accordance with previous reports on the plant secretome ( Agrawal et al., 2010; Shinano et al., 2011; Gupta et al., 2011; Krause et al., 2013; Lum et al., 2014; Li et al., 2012; Ali et al., 2014; Ves-Urai et al., 2021). GSS-resident BET domain-containing proteins (S304, S489) have previously been observed in the chickpea secretome ( Gupta et al., 2015).
2.5. Distinct and shared features of GSS
Prior to this report, several research groups have identified proteins secreted from crop and model species using in vitro setup of SCCs (Supplementary Table S2). We manually curated a database of the in vitro secretome of plants and obtained a set of 5410 sequences. These in turn yielded a set of 3276 nonredundant sequences, which were employed for further downstream comparisons. L. sativus shared ~23% of the sequences with previous reports ( Fig. 3D View Fig ). The unique grasspea secretome sequences were subsequently queried for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, which revealed 23 signal transduction pathways, 7 of which were specific to phytohormone signaling. A species-specific comparison of grasspea was performed with Arabidopsis (326 proteins), chickpea (462 proteins), sorghum (287 proteins) and rice (315 proteins) (Supplementary Fig. S3 View Fig ; grouped from reports in Supplementary Tables S2A and S2B; sequence IDs provided in Supplementary Table S3A). Chickpea, being a leguminous crop, shared the maximum similarity with grasspea (72), highlighting the evolutionary conserved patterns of gene expression in plant families. However, chickpea and grasspea shared a miniscule number of secreted proteins with other plant families. Unique sequences reported for Arabidopsis (174), chickpea (294), sorghum (152) and rice (176), highlight the significance of crop-specific organelle proteomes of plants.
Next, we performed systematic analysis of GSS constituents compared to those of dicots, monocots, and lower plants ( Fig. 3D View Fig ; grouped from reports in Supplementary Table S2; sequence IDs provided in Supplementary Table S3A). As expected, GSS shared maximum proteins with dicots (41), followed by monocots (29) and nonvascular plants (17). The 39 proteins shared by all 4 plant groups included GAPDH, dihydrolipoyl dehydrogenase 1, peroxidase 72, cysteine proteinase RD21A, 5-methyltetrahydropteroyltriglutamate homocysteine methyltransferase 1, pectinesterase/pectinesterase inhibitor 18 and glucan endo-1,3-beta-glucosidase, among others. Grasspea exhibited a unique set of sequences (561), establishing it as a suitable model system to investigate legume biology.
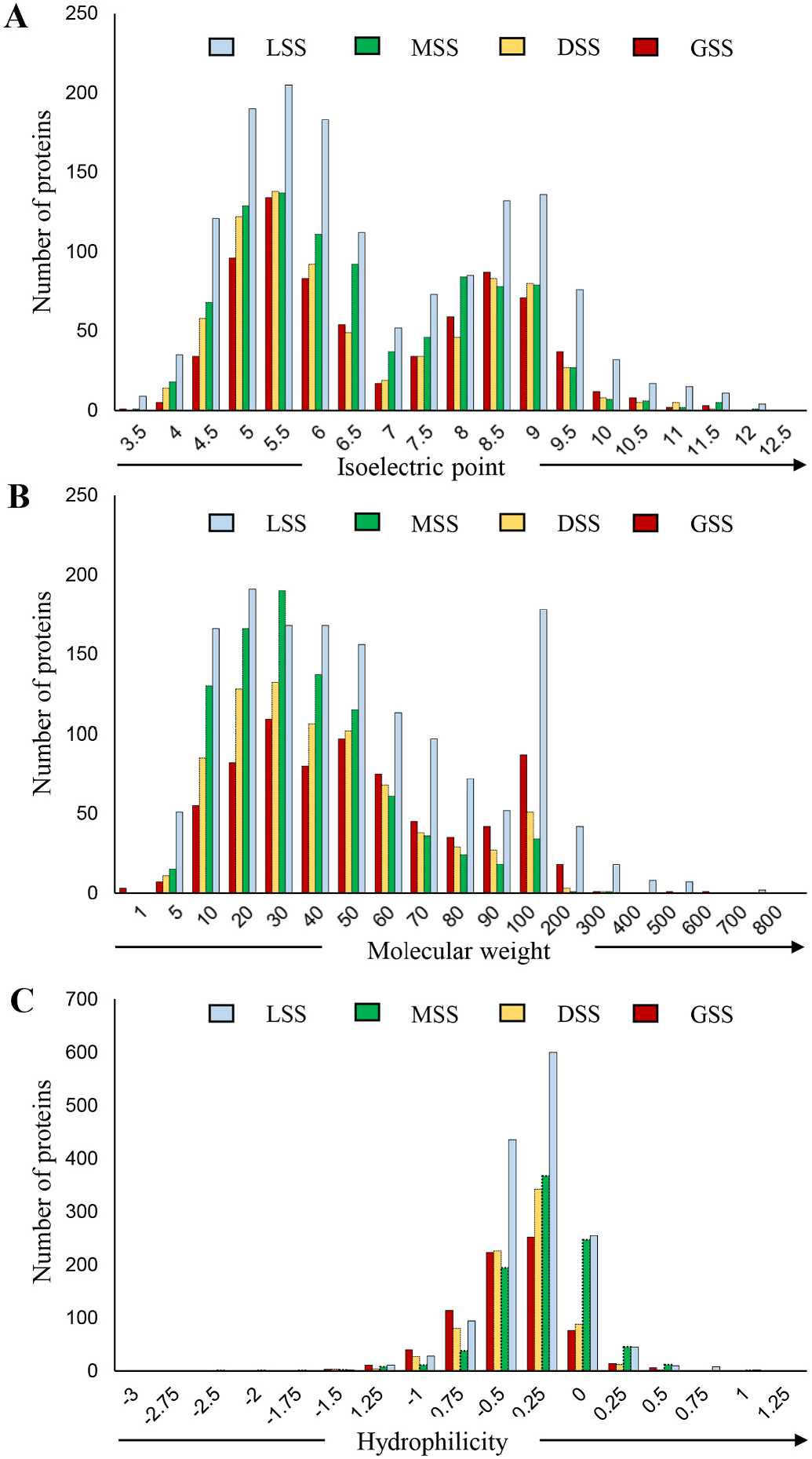
The examination of GSS revealed marked similarity to that of nonvascular plants, monocots, and dicots in the context of physicochemical properties. The isoelectric points of the secreted proteins exhibited an extremely broad range, from 3.5 to 11.5 ( Fig. 4A View Fig ). Similarly, the molecular weights of GSS constituents were as low as 1 kDa and as high as 500 kDa ( Fig. 4B View Fig ). Moreover, GSS proteins were mildly hydrophobic overall, ranging from 1.25 to 0.75 ( Fig. 4C View Fig ). The physicochemical properties of GSS proteins are generally indicative of a broad functional range of secretomes. The secretome biomarkers are family specific, as exemplified by the unique proteins identified using IVS approaches of monocots, dicots and nonvascular plants. While the characteristically low isoelectric points of GSS proteins might be related to their easy secretion into the extracellular space ( Byun et al., 2017), the higher isoelectric points might contribute to a specific functional advantage for the plant secretome. Notably, the neutral to slightly alkaline pI proteins (7–9) are relatively more abundant in monocots and non-vascular plants as compared to the dicots. The high molecular weight plant secretome components might be related to the function of anchorage as integral components of the ECM ( Marinkovich et al., 1992). Also, the high molecular weight proteins may be selectively exported to the extracellular space through UPS, either as constituents of exosomes or multi-vesicular bodies or other Golgi bypass routes. The low molecular weight GSS constituents encompass the essential signaling components in developing regions of plants ( Stahl et al., 1999; Lee et al., 2003; Yokota et al., 2004; Hu et al., 2021) as well as plant immunity modules ( Van de Velde et al., 2010; Yu et al., 2021). The mildly hydrophobic nature of GSS proteins might be due to their transient association with plant membranes and even biomolecules of hydrophobic nature or proteins with hydrophobic cores.
2.6. GSS functional annotation
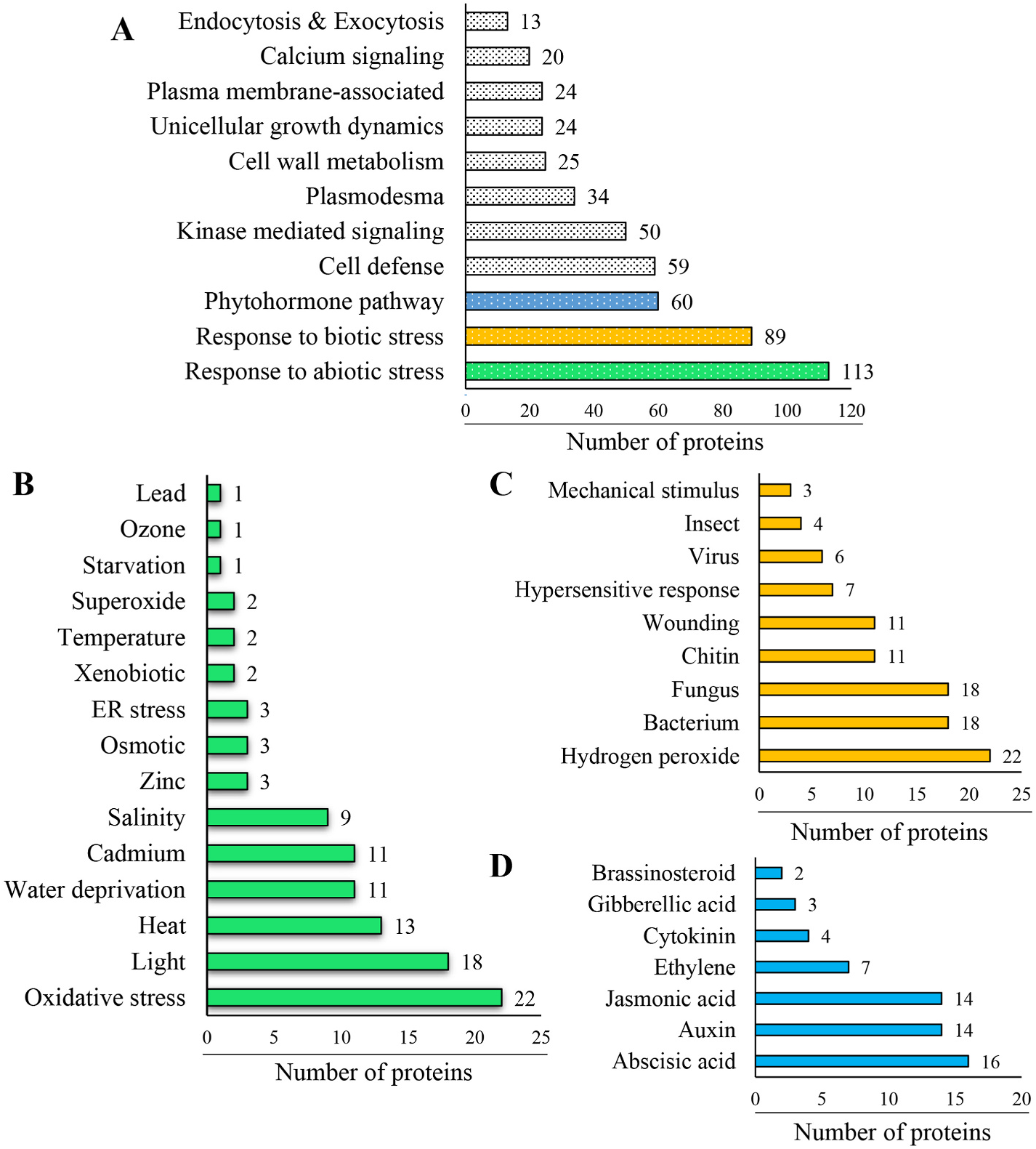
The GSS was sequentially subjected to Gene Ontology (GO) annotation, wherein each protein possesses multiple GO terms. The biological processes were dominated by metabolic, biosynthetic and transportrelated proteins ( Fig. 5A View Fig ). Interestingly, 59 proteins were found to be associated with defense response, while 113 with abiotic stress response. The cellular component category revealed an abundance of membrane-associated proteins ( Fig. 5A View Fig , Supplementary Table S1). The three most abundant GSS functional categories were abiotic stress response ( Fig. 5B View Fig ), biotic stress response ( Fig. 5C View Fig ) and phytohormone signaling ( Fig. 5D View Fig ). Molecular function was mostly related to nucleotide and ion binding, and a significant number of proteins were associated with transferase activity, particularly phosphorus-containing groups. We identified rare proteins associated with exocytosis as well as endocytosis. Since the secretome constituents overlap with those of the cell wall, we identified several primary and secondary cell wall biosynthesis and modifying enzymes in addition to cuticle synthesis-related proteins (eceriferum 3 isoforms). Plant developmental proteins critical for root (12 proteins), leaf (5), seed (5), embryo (12) and flower (16) development were well represented in the secretome. Since totipotent cells were used for generating the in vitro secretome, it was comprised of several crucial proteins (24) related to cell growth, shape, and division. The cell–cell signaling-associated secreted proteins could be classified into transporters, symporters, antiporters, membrane-associated receptors and signaling proteins and plasmodesmata-inhabited proteins.
2.7. Subcellular localization of target proteins
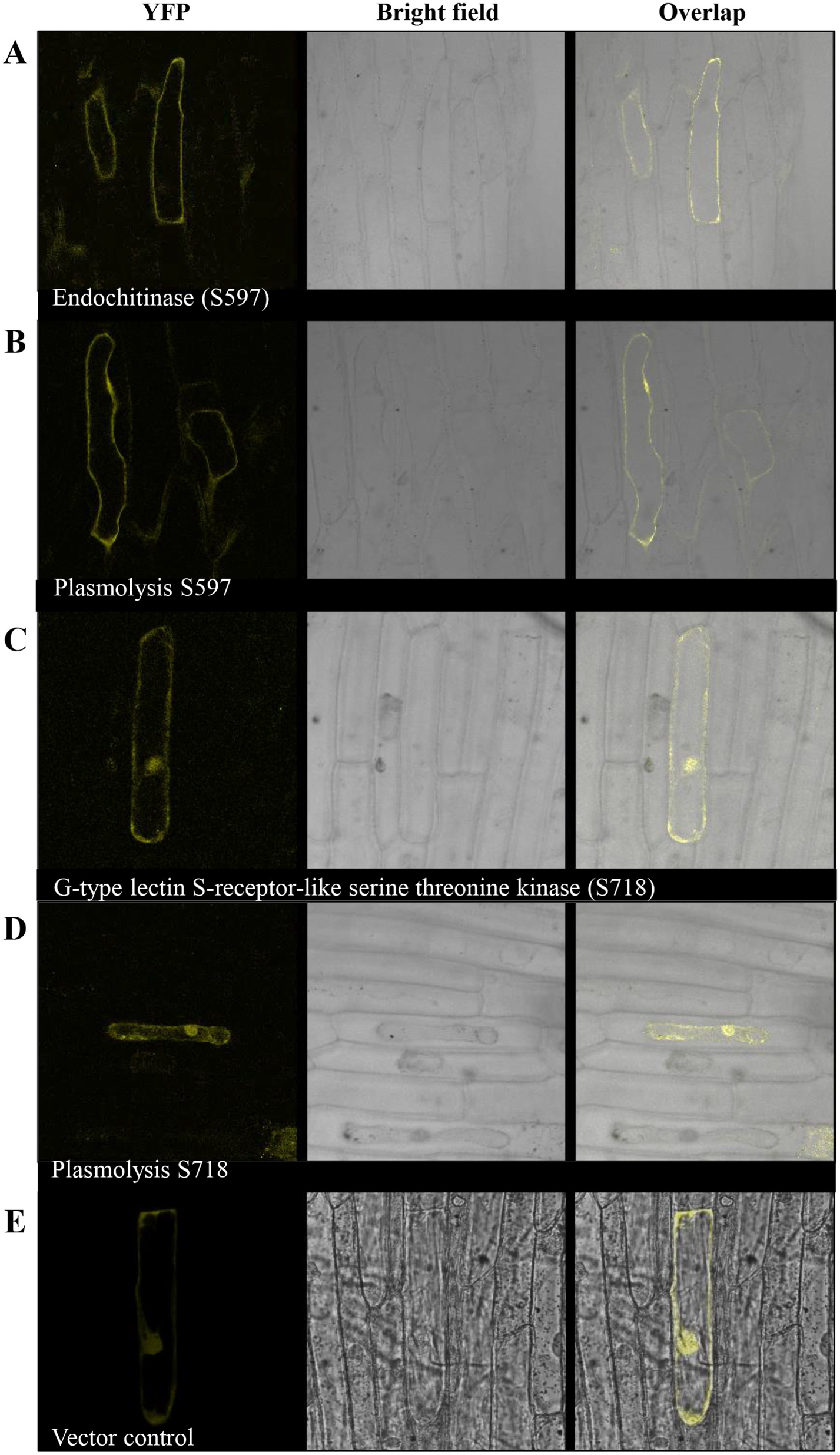
We selected two proteins, endochitinase (S597) and G-type lectin Sreceptor-like serine threonine kinase (S718), predicted to be functional constituents of GSS, to investigate the in planta localization. Endochitinases are integral components of the plant secretome due to their function as antifungal proteins ( Agrawal et al., 2010; Krause et al., 2013; Yadav et al., 2015). On the other hand, G-type lectin S-receptor-like serine threonine kinases are crucial for cell surface signaling and have been reported to be plasma membrane-associated proteins ( Khoza, 2017; von Aderkas et al., 2018; Schellenberger et al., 2019). The localization of target proteins was restricted to the cell surface, as revealed by plasmolyzed protoplasts, especially for endochitinase ( Fig. 6A and B View Fig ). However, in the case of G-type lectin S-receptor-like serine threonine kinase, diffuse expression of the tagged protein was also observed in the cytoplasm, in addition to the cell surface ( Fig. 6C and D View Fig ).
3. Conclusions
In the present study, we attempted to decode the secretome of an orphan legume species known for its unique agricultural traits. Cell-to-cell communication, signaling and extracellular space modifying enzymes were abundantly represented in the GSS. Our findings suggest that totipotent SCCs have the potential to redifferentiate into tissues and organs, depending on the environmental cues. We observed that L. sativus cells are capable of secreting proteins related to key developmental cues and phytohormone-associated pathways. The evolutionary conservation of secretory proteins is evident in the species-specific comparison, more so in the case of plant families. These proteins can be established as “plant secretome markers”. The transient localization analysis of key signaling proteins validated their secretion from the cell surface in nature. Overall, L. sativus exhibited both shared and distinct components of the cell surface, some of which were hitherto undiscovered. Conclusively, the proteome landscape portrayed not only the plant-specific secreted protein markers but also novel constituents of the grasspea secretome.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |