Certhiasomus, Derryberry, Elizabeth, Claramunt, Santiago, Chesser, Terry, Aleixo, Alexandre, Cracraft, Joel, Moyle, Robert G. & Brumfield, Robb T., 2010
|
publication ID |
https://doi.org/ 10.5281/zenodo.293912 |
|
DOI |
https://doi.org/10.5281/zenodo.5667809 |
|
persistent identifier |
https://treatment.plazi.org/id/7832E513-161F-FFD7-12C5-FF04FD418693 |
|
treatment provided by |
Plazi |
|
scientific name |
Certhiasomus |
| status |
gen. nov. |
Certhiasomus , new genus
Type species. Sittasomus stictolaemus Pelzeln 1868 .
Included taxa. Certhiasomus s. stictolaemus ( Pelzeln 1868) , Certhiasomus stictolaemus secundus ( Hellmayr 1904) , Certhiasomus stictolaemus clarior ( Zimmer 1929) .
Diagnosis. Small woodcreeper (13–22 g, Marantz et al. 2003) with relatively long tail and slender bill. Certhiasomus differs from Dendrocincla and all “strong billed woodcreeper genera ( Drymotoxeres , Drymornis , Nasica , Dendrexetastes , Hylexetastes , Xiphocolaptes , Dendrocolaptes , Xiphorhynchus , Lepidocolaptes and Campylorhamphus ; Feduccia 1973) by a combination of small size, operculated nostrils, and rectrices with long, strongly decurved protruding rachises covered with short stiff barbs. Distinguished from Glyphorynchus by its longer non-wedge-shaped bill, and from Sittasomus by its light throat markings and more pronounced tail graduation. Distinguished from Deconychura by its smaller size, absence of an apical bill hook, nearly unstreaked crown, and extensive rufous coloration on rump (restricted to upper tail coverts in Deconychura ). Myologically, it is distinguished from Deconychura by the iliotibialis lateralis pars postacetabularis muscle, the caudal margin of which arises even with the caudal edge of the iliofibularis muscle in Certhiasomus , and cranial to it in Deconychura ( Raikow 1994) .
Etymology. From the Greek certhia (treecreeper) and soma (body), referring to the morphological similarities between this small woodcreeper and the treecreepers Certhiidae . The construction of the name parallels that of the closely related Sittasomus , the genus in which C. stictolaemus was originally described. The name is masculine in gender.
Molecular analyses. Our preliminary molecular analysis of all dendrocolaptid species found that Certhiasomus is sister to a clade composed of Dendrocincla , Deconychura , and Sittasomus . To demonstrate that Deconychura and Certhiasomus are not sister genera, we present an analysis of a subset of taxa from this larger study. We included in the analysis individuals of Certhiasomus stictolaemus , Deconychura longicauda , Dendrocincla turdina ( Lichtenstein) (Plain-winged Woodcreeper), Sittasomus griseicapillus , Drymornis bridgesii (Eyton) (Scimitar-billed Woodcreeper), Glyphorynchus spirurus (Vieillot) (Wedge-billed Woodcreeper), Dendroplex picus (Gmelin) (Straight-billed Woodcreeper), and Lepidocolaptes lacrymiger (Des Murs) (Montane Woodcreeper) ( Table 1 View TABLE 1 ). We included six individuals of C. stictolaemus , two for each of the three described subspecies: C. s. stictolaemus , C. s. secundus , and C. s. clarior . We sequenced multiple individuals of D. longicauda , including one individual of the typica group from Central America. Marantz et al. (2003) suggested that this group might be more closely related to C. stictolaemus than to the Amazonian forms of D. longicauda . To root the tree we used as outgroups Pygarrhichas albogularis (King) (Whitethroated Treerunner, Furnariidae ), Myrmothera simplex (Salvin & Godman) (Brown-breasted Antpitta, Grallariidae ) and Formicarius colma Boddaert (Rufous-capped Antthrush, Formicariidae ).
Total DNA was extracted from 25 mg of pectoral muscle using the Qiagen DNeasy kit, following the manufacturer's protocol. Following methods described in Chesser et al. (2007), we amplified and sequenced the mitochondrial genes ND3 and CO2 and the autosomal nuclear intron BF7. To amplify CO2, we used two primers newly designed for furnariids, NF3 ( Sanín et al. 2009) and SCTRCOII (Claramunt et al. in press). Following the same methods, we amplified and sequenced an additional mitochondrial gene (ND2) for at least one individual per species using the primers H6313 ( Johnson & Sorenson 1998) and L5215 ( Hackett 1996). For at least one individual per genus, two additional nuclear protein-coding genes (RAG1 and RAG2) were used; all sequences of these genes were taken from Moyle et al. (2009). Following alignment and the exclusion of unique inserts from the BF7 sequences, the six-gene concatenated dataset consisted of 6,990 base pairs.
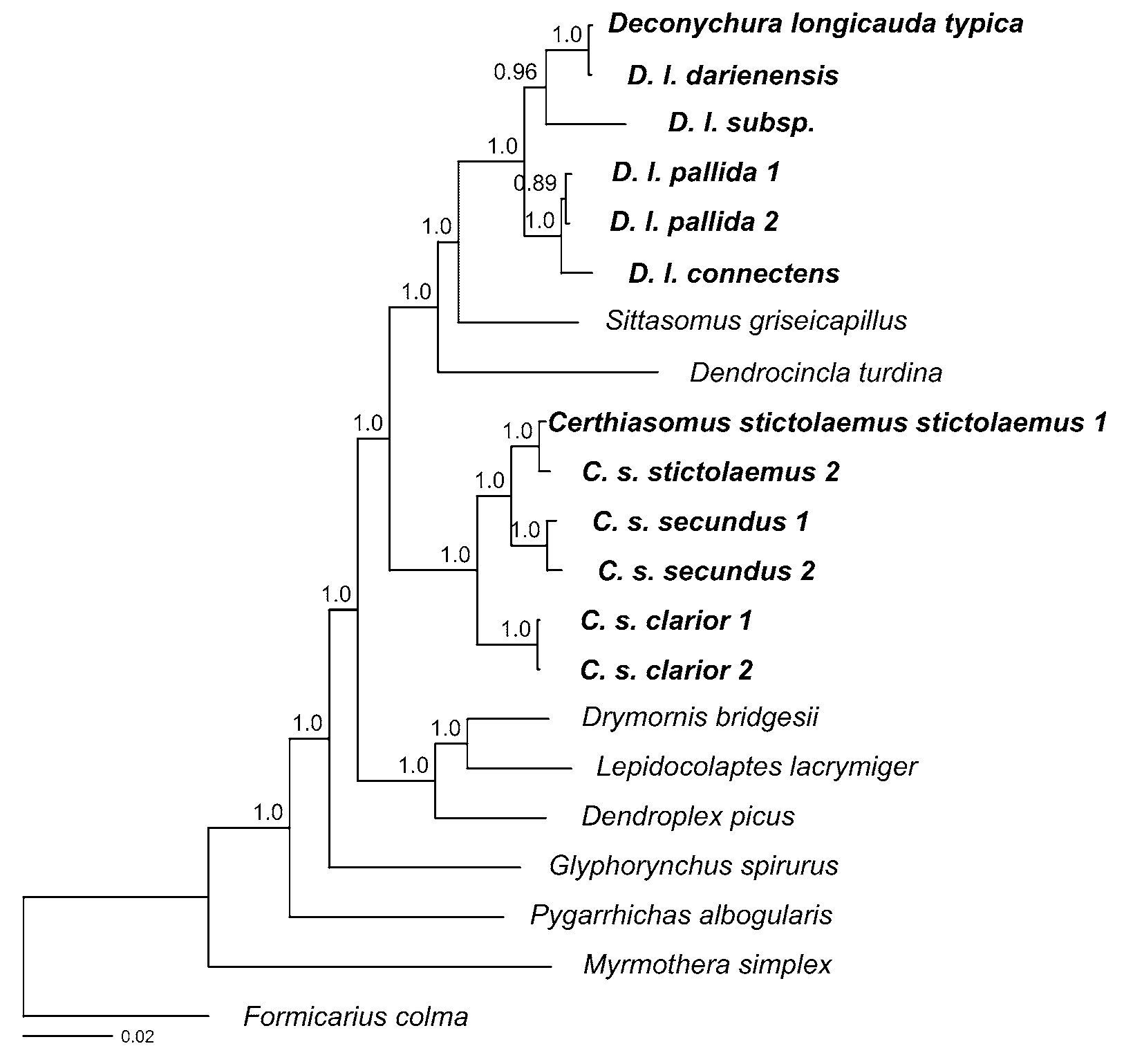
In model-based phylogenetic inference, there is a trade-off between capturing the complexity of the nucleotide substitution process and the risk of over-parameterizing the models ( Posada & Buckley 2004; Sullivan & Joyce 2005; McGuire et al. 2007). To determine the optimal number of partitions, we tested six different partitioning regimes, ranging from unpartitioned to a maximum of sixteen different partitions (a different model for each codon position of each coding gene and the nuclear intron). Partitioned datasets were examined by performing maximum likelihood (ML) analyses using RAxML 7.0.4 on the Cipres Portal V 1.5 (www.phylo.org/sub_sections/portal/). RAxML implements the GTR+Γ model of nucleotide substitution (with or without invariants). Using the resultant likelihoods we calculated values of the Akaike Information Criterion (AIC, Sullivan & Joyce 2005) for each partitioning regime. We also investigated whether the addition of a proportion of invariant sites improved the model. We identified the GTR+ Γ+I model and a fully partitioned dataset (16 partitions) as the best model and partitioning regime. We then used RAxML to evaluate nodal support in the resulting maximum-likelihood tree by performing 1000 bootstrap replicates ( Stamatakis et al. 2008). The analysis resulted in a single maximum-likelihood tree (log L = 20923.4) with high bootstrap support for most relationships ( Fig. 1 View FIGURE 1. A ).
Tissue collections: LSUMNS—Louisiana State University Museum of Natural Science, Baton Rouge; AMNH—American Museum of Natural History, New York City; FMNH—Field Museum of Natural History, Chicago; MPEG—Museu Paraense Emílio Goeldi, Belém, Brazil
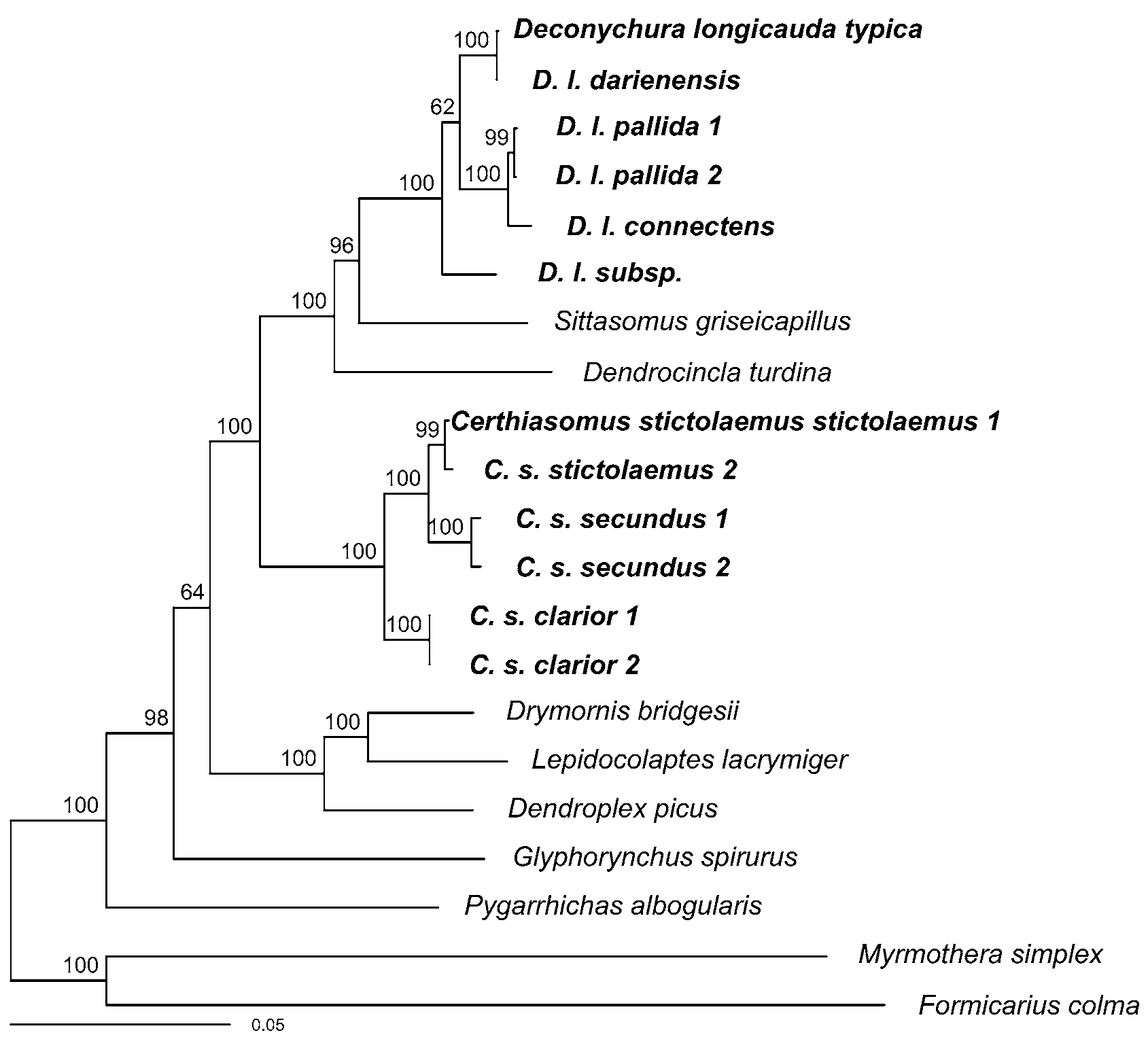
As a second method of phylogenetic inference, we performed a Bayesian analysis as implemented in MRBAYES Ve r. 3.0 β3 ( Huelsenbeck & Ronquist 2001; Altekar et al. 2004) on the Cornell cluster (http:// cbsuapps.tc.cornell.edu/mrbayes.aspx). We used the same partitioning strategy as in the likelihood analysis, but again used model selection to determine the best substitution model for each partition. For each partition, using the tree obtained in the maximum-likelihood analysis, we used PAUP ( Swofford 2003) to obtain likelihood values for all substitution models featured in Modeltest 3.7 ( Posada & Crandall 1998) and then calculated values of the Bayesian Information Criterion (BIC, Posada and Buckley 2004; Sullivan & Joyce 2005) for each. BIC identified the GTR+Γ+I model as the best model for the majority of the partitions, and the HKY+Γ+I model as the best model for the first and second codon positions of RAG 1 and all three codon positions of RAG 2. The Bayesian posterior probability density was estimated by Metropolis-coupled Markov chain Monte Carlo, with four incrementally heated chains run for 25 million generations (sampled every 2500). All chains reached stationarity, and all parameters had good ESS values (>200). The analysis resulted in a topology almost identical to the best maximum-likelihood tree with high posterior support for most relationships ( Fig. 2 View FIGURE 2 ).
In both analyses, all individuals of D. longicauda , including D. l. typica , formed a strongly supported clade (bootstrap support = 100%, posterior probability = 1.0) that was sister to Sittasomus and Dendrocincla . The ( D. longicauda , Sittasomus , Dendrocincla ) clade was sister to Certhiasomus with 100% bootstrap support and a posterior probability of 1.0. These results clearly demonstrate that Certhiasomus and Deconychura do not form a clade.
Phenotypic analyses. Although phenotypic differences among Sittasomus , Deconychura and Certhiasomus could be considered slight and not of generic significance (see diagnosis), the genus Dendrocincla can be distinguished by several unique characters, including the structure of their rectrices, myological synapomorphies ( Raikow 1994), and foraging behavior atypical of dendrocolaptids ( Skutch 1969; Willis 1972; Marantz et al. 2003). Therefore, uniting all these genera in a single genus (the alternative to describing a new genus) would result in a taxon that is excessively heterogeneous relative to taxa of similar rank in the Dendrocolaptidae . Traditionally, problems like this have been treated subjectively in taxonomic work. Here, we have approached the problem of taxon heterogeneity using a quantitative approach by means of a morphometric analysis of total variance (Claramunt et al. in press).
The following measurements were taken for at least three museum specimens of each of the 50 currently recognized species of Dendrocolaptidae : wing length to the longest primary, wing length to the tenth (most distal) primary, wing length to the first secondary feather, tail maximum (to the longest rectrix) and minimum (to the shortest rectrix) length, width of the innermost rectrix, bill length from naris, bill width and depth at the anterior edge of naris, tarsus length, and hallux length with claw (for further details, see Claramunt et al. in press). Variables were log-transformed ( Gingerich 2000). To separate the effect of body size from size-free shape variation, we used Mosimann's approach ( Mosimann 1970; Mosimann & James 1979). We estimated the isometric size of each species as the mean of the log-transformed variables. We then calculated size-free variables by subtracting the log(size) of the species from each variable.
Morphometric heterogeneity was quantified as the total variance ( Van Valen 1974). We computed the total variance for all non-monotypic dendrocolaptid genera plus a hypothetical genus encompassing species of Dendrocincla , Sittasomus , Deconychura , and Certhiasomus . We also estimated the separate contribution of size and shape to the total variance ( Darroch & Mosimann 1985). Comparison of size and shape heterogeneity indicated that the morphometric variance of the hypothetical, more inclusive genus is more than twice that of the next most heterogeneous dendrocolaptid genus (Table 2). These data support our decision to create a new genus ( Certhiasomus ) rather than create an extremely heterogeneous genus by combining Dendrocincla , Sittasomus and Deconychura into one.
TABLE. 2. Results of the morphometric heterogeneity analysis.
Taxon Total Variance Size variance Shape Variance Dendrocincla + Deconychura 1 + Sittasomus 0.379 0.302 0.077 Dendrocincla 0.170 0.138 0.033 Xiphorhynchus 0.169 0.140 0.030 Campylorhamphus 2 0.064 0.028 0.036 Lepidocolaptes 0.061 0.034 0.027 Xiphocolaptes 0.045 0.026 0.020 Dendrocolaptes 0.030 0.010 0.020 Hylexetastes 0.021 0.005 0.016 Dendroplex 0.021 0.012 0.009
1including both D. longicauda and D. stictolaema . 2excluding C. pucherani (Claramunt et al. in press).
TABLE 1. Tissue samples used in the genetic analysis.
| Taxon | Museum | Sample ID | Locality |
|---|---|---|---|
| Deconychura longicauda typica | LSUMNS | B26585 | PANAMA: prov. Colón; Río Agua Salud. |
| D. longicauda darienensis | LSUMNS | B2084 | PANAMA: prov. Darién; Cana. |
| D. longicauda pallida 1 | LSUMNS | B905 | BOLIVIA: depto. Pando; Río Beni, 600 m. |
| D. longicauda pallida 2 | LSUMNS | B4753 | PERU: depto. Loreto; S Río Amazonas, 100 m. |
| D. longicauda (subsp. uncertain) | LSUMNS | B27966 | PERU: depto. Loreto; Contamana, 1450 m. |
| Certhiasomus stictolaemus stictolaemus 1 | MPEG | 57571 | BRAZIL: Amazonas; Manicor, Rodovia do Estanho, km 126. |
| C. stictolaemus stictolaemus 2 | MPEG | 58684 | BRAZIL: Amazonas; Município de Humait, T. Indígena Parintintin, Aldeia Traira-Choror. |
| C. stictolaemus secundus 1 | LSUMNS | B27420 | PERU: depto. Loreto; Contamana, 200 m. |
| C. stictolaemus secundus 2 | LSUMNS | B2532 | PERU: depto. Loreto; N Río Napo, 350 m. |
| C. stictolaemus clarior 1 | MPEG | 65378 | BRAZIL: Pará; Alenquer, ESEC Grão-Par. |
| C. stictolaemus clarior 2 | MPEG | A07970 | BRAZIL: Pará; Município de Almerim, Monte Dourado, Reserva de Pacanari. |
| Sittasomus griseicapillus | AMNH | DOT 8415 | MEXICO: Hidalgo; Molango. |
| Dendrocincla turdina | LSUMNS | B250 | PARAGUAY: depto. Caazap; Cor. de Caaguaz, 7.5 km E. San Carlos, 250m. |
| Glyphorynchus spirurus | AMNH | DOT 4274 | VENEZUELA: Amazonas; Sierra de Tapirapeco, Cerro Tamacuari, 1270m. |
| Drymornis bridgesii | LSUMNS | B25799 | PARAGUAY: depto. Alto Paraguay; Madrejón, 200m. |
| Dendroplex picus | FMNH | 334433 | BOLIVIA: depto. El Beni; Laguna Suarez, 5km SW Trinidad, 230m. |
| Lepidocoplates lacrymiger | AMNH | DOT 7051 | BOLIVIA: depto. La Paz; Parque Nacional Apolobamba, 2600m. |
| Pygarrhichas albogularis | AMNH | DOT 9930 | ARGENTINA: depto. Bariloche; Río Negro. |
| Formicarius colma | AMNH | DOT 12722 | VENEZUELA: Amazonas; Cerro de la Neblina Base Camp, Río Baria. |
| Myrmothera simplex | AMNH | DOT 4270 | VENEZUELA: Amazonas; Sierra de Tapirapeco, Cerro Tamacuari, 1270m. |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.