Unio tumidiformis, Castro, 1885
|
publication ID |
https://doi.org/ 10.1093/zoolinnean/zlx039 |
|
DOI |
https://doi.org/10.5281/zenodo.4725735 |
|
persistent identifier |
https://treatment.plazi.org/id/744587AC-FFCC-D525-9EB4-513AFBEF04D1 |
|
treatment provided by |
Carolina |
|
scientific name |
Unio tumidiformis |
| status |
|
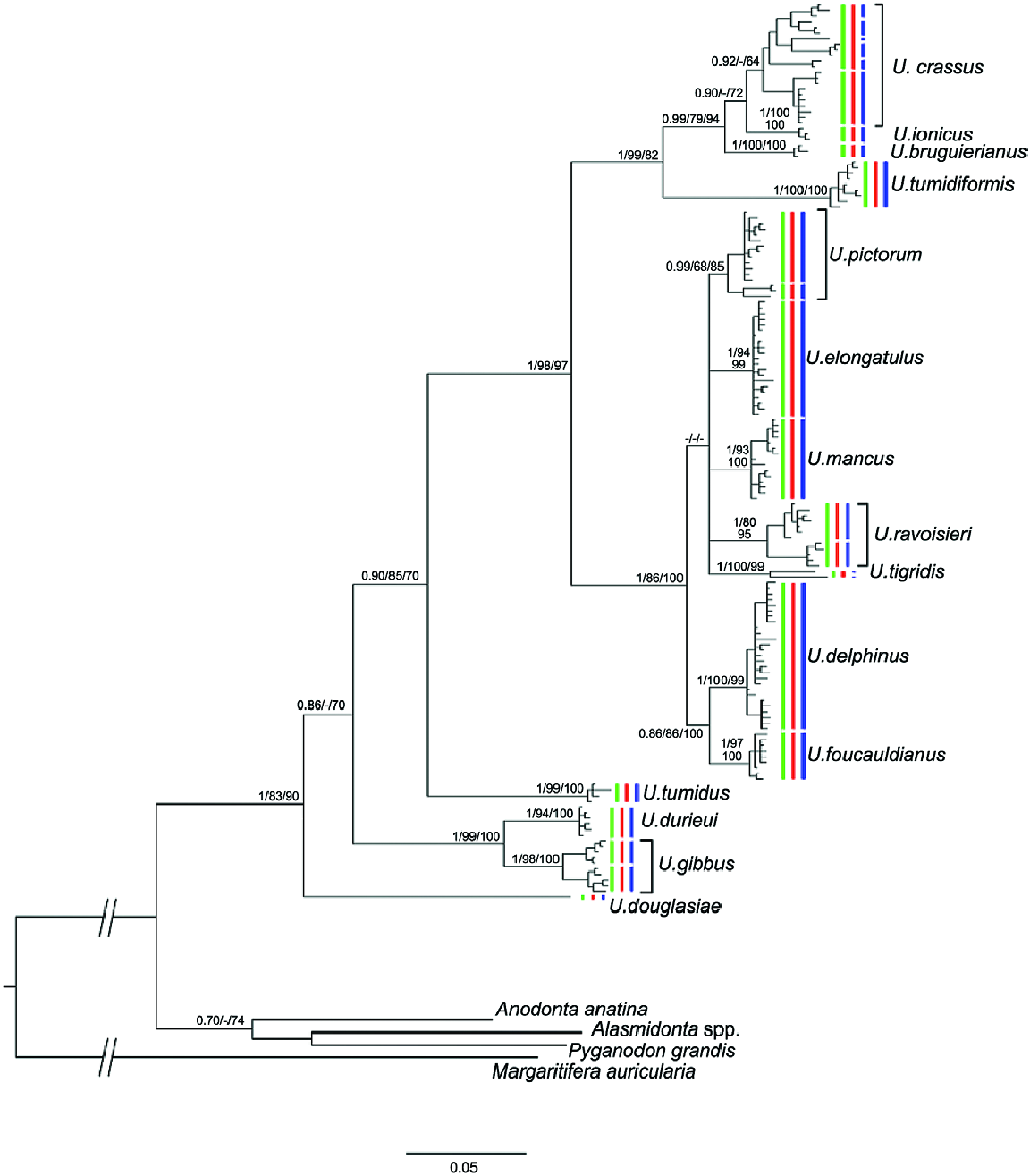
The U. tumidiformis + [U. bruguierianus + (U. ionicus + U. crassus)] lineage
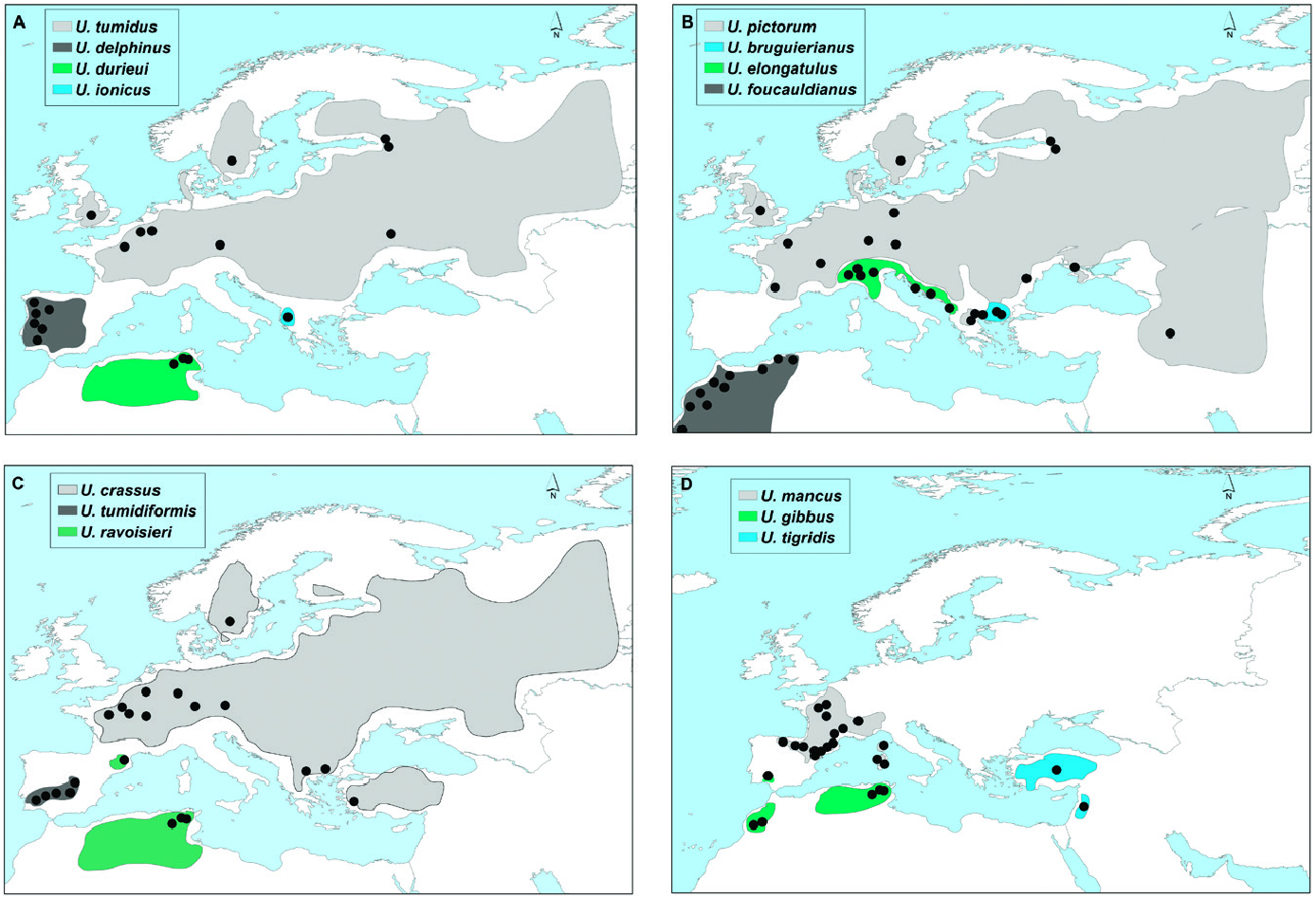
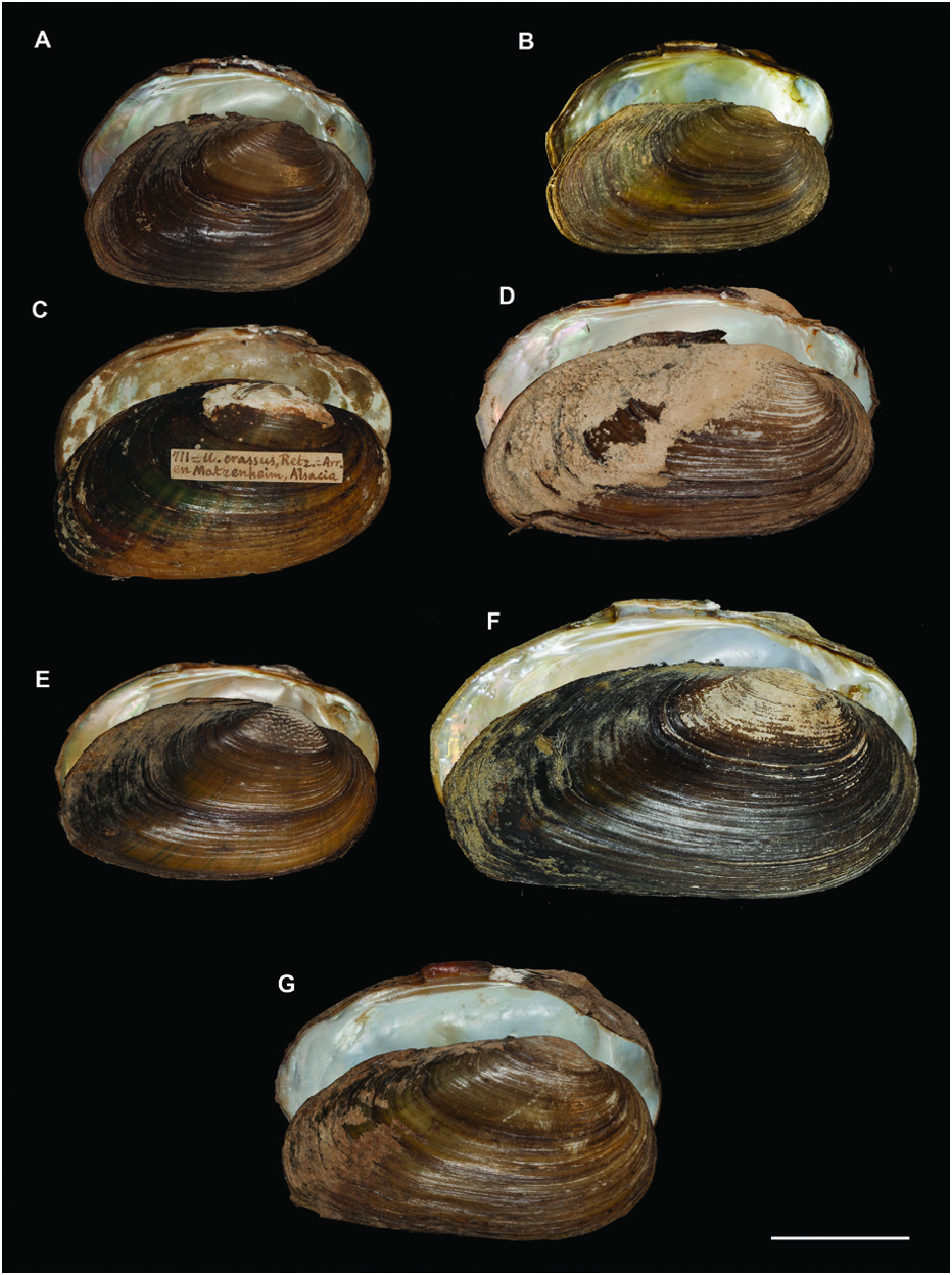
This lineage included two monophyletic groups with a COI genetic distance greater than 7.85%: a western group that included the endemic Iberian species U. tumidiformis ( Fig. 2 View Figure 2 ) and a second group (supported by a posterior probability of 0.99 and bootstrap values of 79 for ML and 94 for MP) comprising three morphospecies with a complex geographic structure ( Fig. 1 View Figure 1 ; Table 1 View Table 1 ).
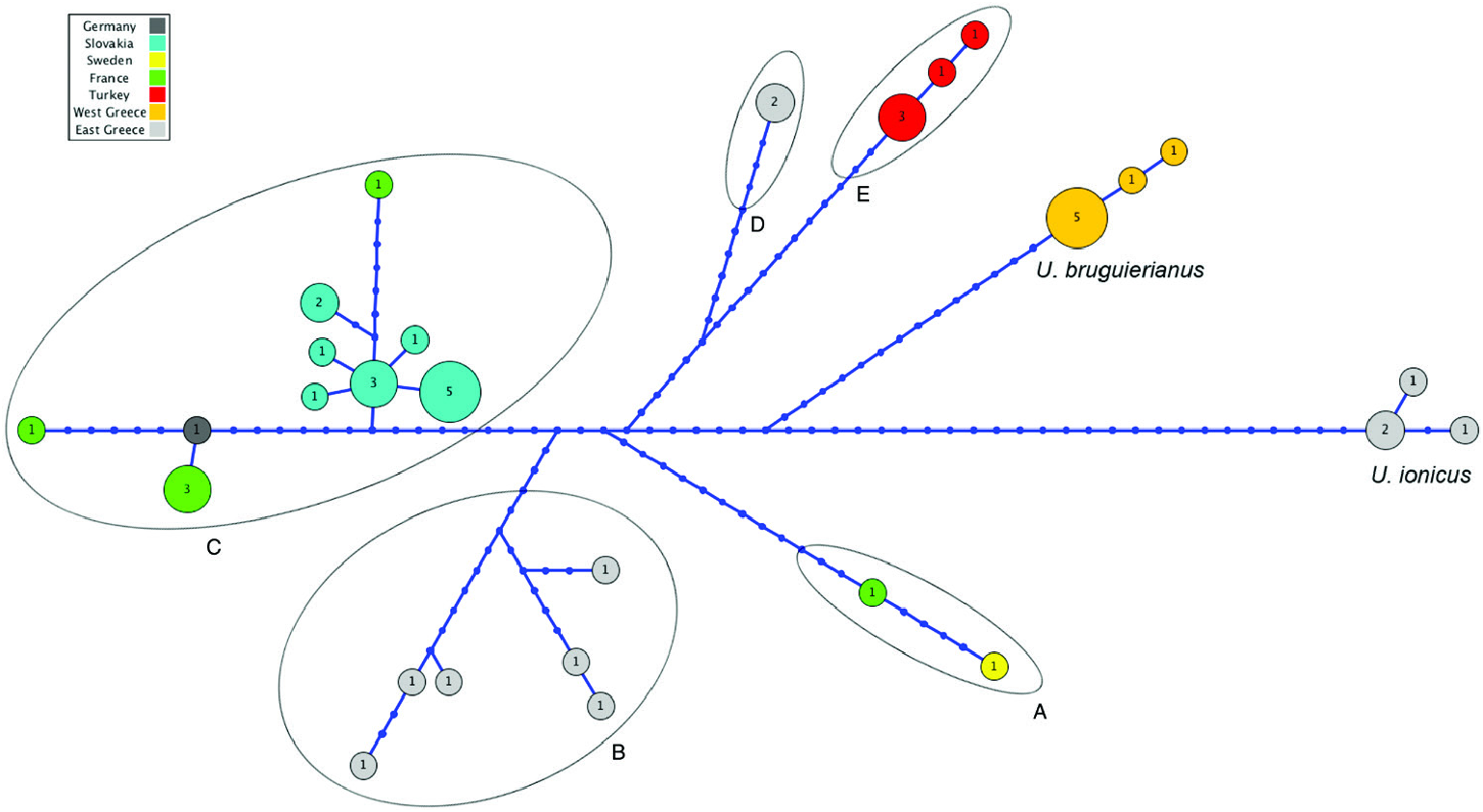
Up to nine haplotypes were found among the samples analysed for U. tumidiformis (Supporting Information, Fig. S10 View Figure 10 ). Two were exclusive to the Portuguese Sado River but were close to some Spanish haplotypes. The most frequent haplotype was found in the Mira and Guadiana rivers.
Within the second group, U. bruguierianus from eastern Greece (the Axios and Pinios rivers) ( Fig. 2 View Figure 2 ) was the sister species of U. ionicus + U. crassus (genetic distance of 4.28–4.45%), although this relationship was not highly supported. Unio ionicus lives in Albania and the Acheloos River (including Lake Lysimacheia) in western Greece ( Fig. 2 View Figure 2 ) and was the sister group of U. crassus (genetic distance of 4.30%). Unio crassus presented five subclades with a minimum divergence of 2.16% ( Table 1 View Table 1 ) and haplotypes separated by at least 19 steps ( Fig. 12 View Figure 12 ). These five subclades included (1) U. crassus courtillierii Hattemann, 1859 from France and one sample from Sweden (both sequences obtained from GenBank); (2) samples from the Sofaditikos (Pinios catchment), Aliakmon/Aliakmonas and Sperchios rivers in eastern Greece; (3) Central European samples from the Rhine, Danube, and Rhône catchments; (4) samples from the Lissos River (eastern Greece); and (5) samples from western Turkey ( Fig. 2 View Figure 2 ). This group had the most complexity, which is also reflected in the bGMYC and M-PTP analyses in which up to six species were revealed.
All U. crassus haplotypes were represented in a unique network ( Fig. 12 View Figure 12 ). No clear biogeographic structure was observed, and some haplotypes belonging to specimens from the same rivers had a high number of substitutions (up to 33). From the right end of Figure 12 View Figure 12 , three haplotypes from eastern Greece were assigned to U. bruguierianus . Three distinct haplotypes from western Greece and Albania were considered to represent U. ionicus . Three haplotypes were found from the Turkish locations analysed, but with a smaller genetic distance compared with the other haplotypes.
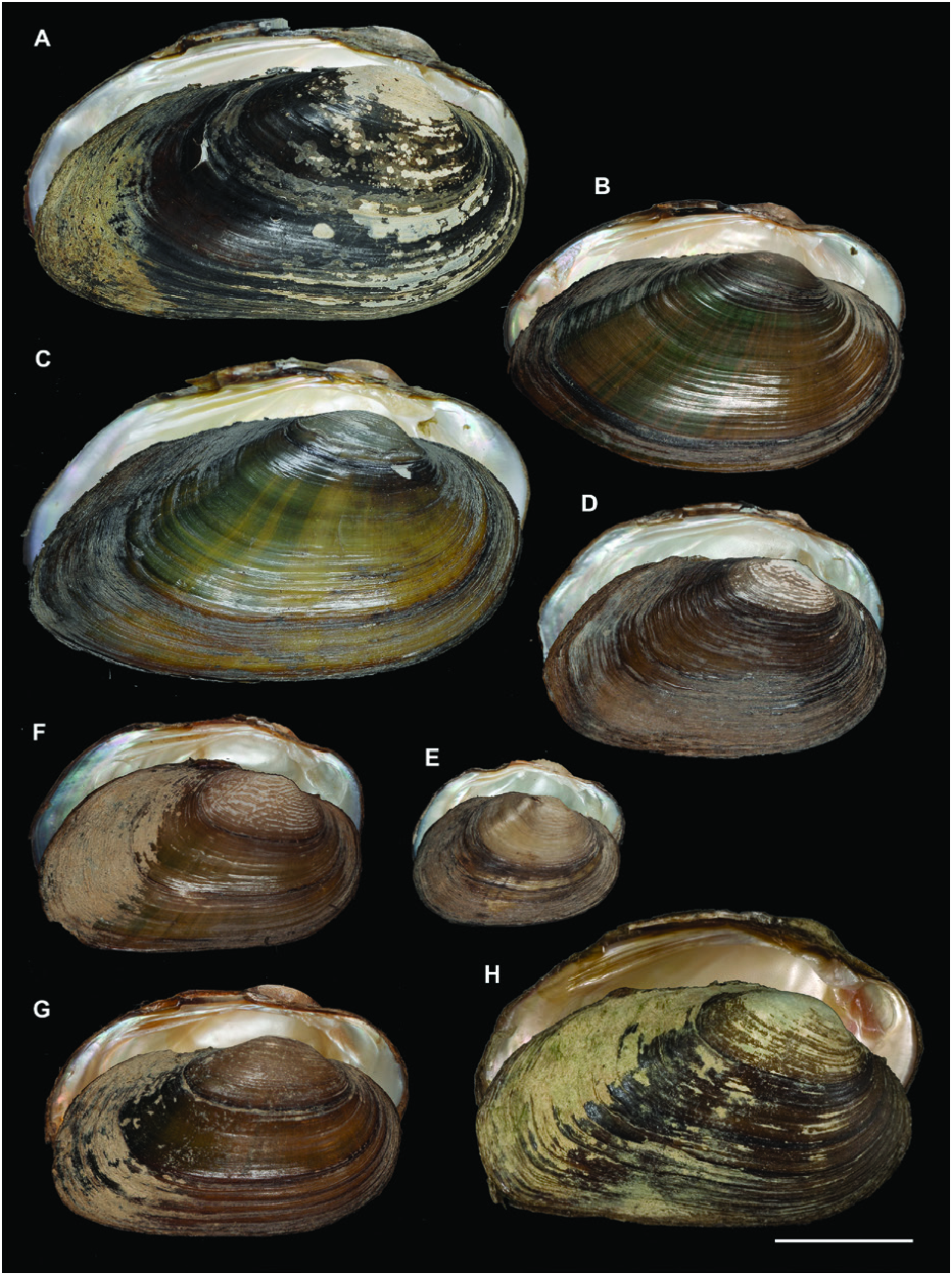
A polymorphic shell shape was observed for U. bruguierianus : some specimens from the Axios and Pinios rivers had a shell shape resembling some pictorum specimens from the Axios River, while other specimens from the Pinios River were identical to some U. crassus specimens from the Lissos River or Turkey ( Fig. 13 View Figure 13 ). This polymorphic shell shape was also observed for U. ionicus and U. crassus ( Figs 13 View Figure 13 , 14 View Figure 14 ). Unio ionicus had an almost identical shell to some U. crassus specimens from the Sofaditikos (Pinios Basin), Sperchios and Lissos rivers in Greece. In contrast, other U. crassus specimens from the Lissos River and from Turkey presented an elongated shell shape. There were also specimens with and without sculpture in the umbo area from the same locality with the same haplotype, such as those from the Lissos River ( Fig. 14 View Figure 14 ).
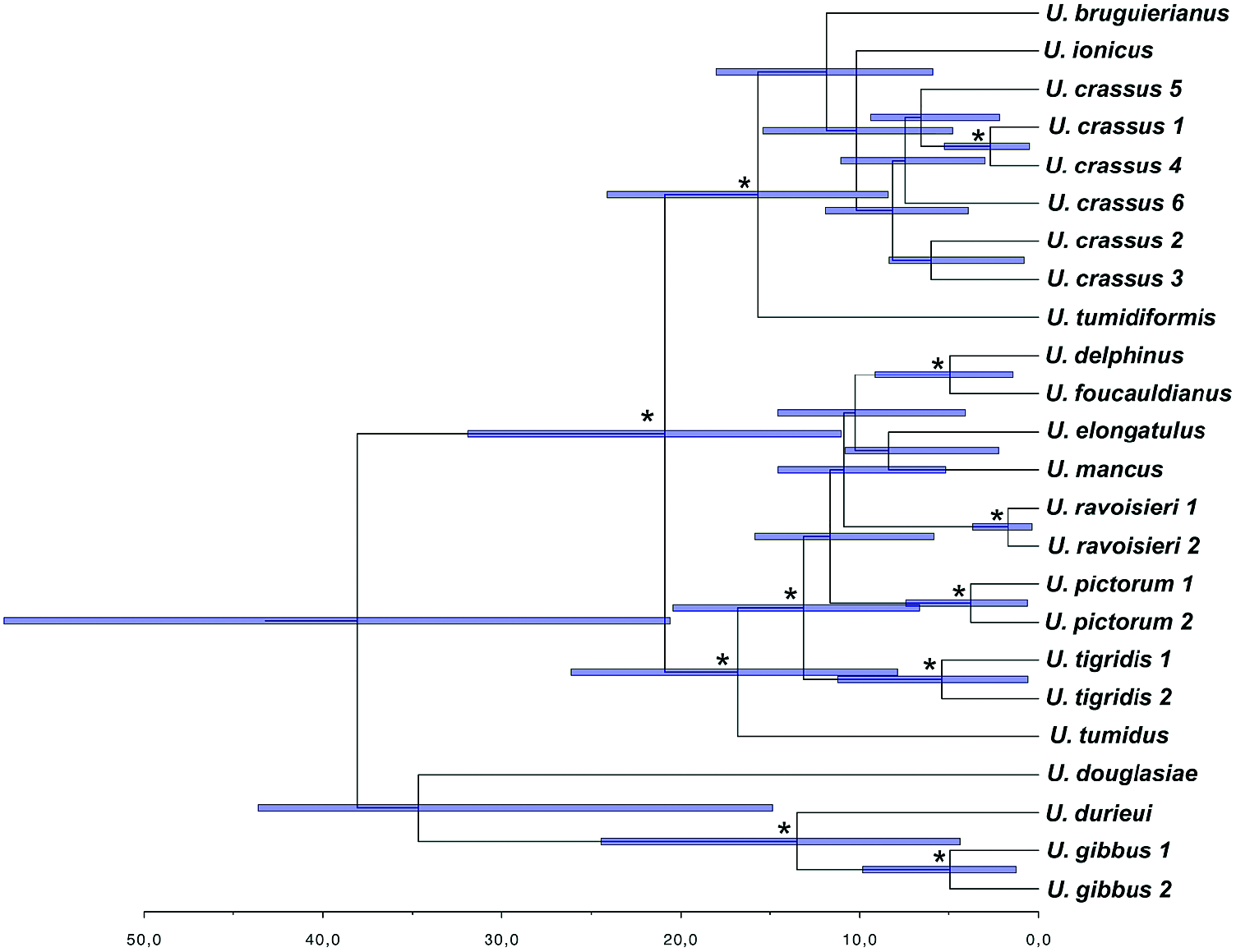
The coalescence-based molecular clock analyses for the species tree resulted in very broad temporal intervals for most of the clades. Although the results have to be interpreted with caution, they provide a broad temporal framework for the diversification of the group. The coalescence reconstruction ( Fig. 15 View Figure 15 ) placed the common ancestor of the Unio clade and the origin of the Western Palaearctic Unio in the Eocene. In fact, the Western Palaearctic species underwent two main cladogenetic events. The first event occurred in the Early Eocene ( U. gibbus , U. durieui split), and the second during the Oligocene, involving the divergence of the most speciose clade. Most of the modern species appeared during the Miocene. The most recent cladogenetic event involved the U. delphinus and U. foucauldianus clade at the end of the Miocene (Messinian). Notably, the reconstructed species tree agrees with the general topology of the concatenated matrix analyses except in the phylogenetic position of U. tumidus . In the species tree, U. tumidus appeared with strong support as sister group of the U. tigridis + [ U. pictorum , U. ravoisieri ( U. mancus + U. elongatulus , U. delphinus + U. foucauldianus )] lineage ( Fig. 15 View Figure 15 ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.