Encarsia heratyi Polaszek
|
publication ID |
https://doi.org/ 10.5281/zenodo.203137 |
|
DOI |
https://doi.org/10.5281/zenodo.6194117 |
|
persistent identifier |
https://treatment.plazi.org/id/591087F3-4360-F928-198F-00E1018FAA12 |
|
treatment provided by |
Plazi |
|
scientific name |
Encarsia heratyi Polaszek |
| status |
sp. nov. |
Encarsia heratyi Polaszek sp.n.
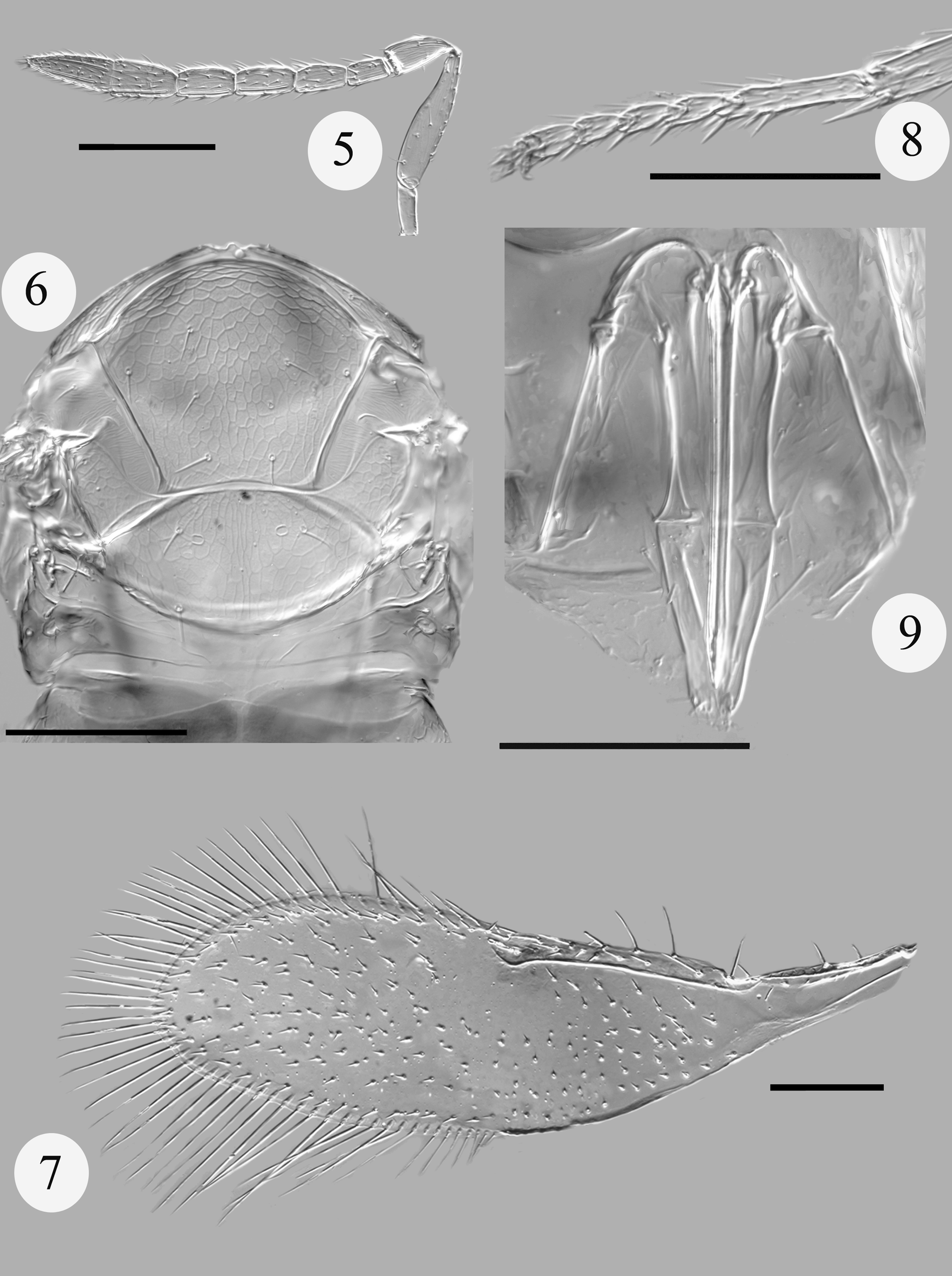
( Figs 5–9 View FIGURES 5 – 9 )
Female (holotype). Colour. Head yellow except brown area present laterally across occiput and vertex with two brown patches behind stemmaticum. Mesosoma yellow except following brown; pronotum, prosternum and anterior half of mesoscutellar midlobe, acropleuron, prepectus, most of ventral mesosoma, and propodeum. Antenna yellow/pale brown except apical two segments (clava) darker brown. Fore wing hyaline, but with a very distinct dark spot at distal end of marginal vein extending to below stigmal vein. Legs yellow. Metasoma largely brown except T3-T4 and T7 paler. Third valvulae entirely yellow.
Morphology. Antennal formula 1,1,4,2 ( Fig. 5 View FIGURES 5 – 9 ). Pedicel longer than F1 (1.6). F1 1.8 times longer than its maximum width, shorter than F2 (0.76) and F3 (0.65). F2 slightly shorter than F3 (0.86). Flagellomeres with the following numbers of sensilla: F1: 0, F2: 0, F3: 2, F4: 2, F5: 3, F6: 2. Stemmaticum and frons with aciculate sculpture, that of stemmaticum approaching a reticulate condition. Mid lobe of mesoscutum ( Fig. 6 View FIGURES 5 – 9 ) with 4 pairs of setae, side lobes with 2 setae each. Scutellar sensilla separated by 5 times maximum width of a sensillum. Distance between bases of anterior pair of scutellar setae very slightly less than that between bases of posterior pair, anterior setae slightly shorter than posterior setae. Fore wing ( Fig. 7 View FIGURES 5 – 9 ) 3.3 times maximum width of disc, with asetose area distal to, and below, stigmal vein. Marginal fringe 0.66 times as long as maximum width of disc. Basal cell with a single seta. Submarginal vein with 2 setae, marginal vein anteriorly with 5 setae. Tarsal formula 5-5-5. Apical spur of mid tibia half as long as the corresponding basitarsus (0.48; Fig. 8 View FIGURES 5 – 9 ). Metasomal terga with following numbers of setae laterally: T1: 0, T2: 1, T3: 1, T4: 1, T5: 2, T6: 0, T7: 4. T5 and T6 also with a pair of setae centrally. Ovipositor ( Fig. 9 View FIGURES 5 – 9 ) as long as mid tibia (or very slightly shorter) and 1.6 times longer than clava. Third valvulae apically rounded, external sides slightly convex, 0.7 times as long as second valvifers.
Variation. F4 with 2–3 sensilla; relative proportions of antennal flagellomeres varying slightly.
Male. Colour. Very similar to that of female, the main difference being an entirely dark metasoma, and darker legs. Fore wing entirely infuscate below marginal vein, infuscation extending to wing base.
Morphology. Structure, sculpture and setation very similar to those of female. Antenna with 8 antennomeres; F6 partially fused. Flagellomeres with the following numbers of sensilla: F1: 5, F2: 4, F3: 4–5, F4: 3–4, F5: 4, F6: 4.
Material examined. Holotype female: U.S.A., California, Riverside, County, Deep Canyon 33º38’N 116º23’W, 25.iii.2009, coll. A. Polaszek on Hyptis emoryi ( BMNH).
Paratypes (same data as holotype): 2 females, 4 males (in BMNH; UCR).
Distribution. U.S.A., California.
Host. Aleuropleurocelus serratus Gill ( Hemiptera : Aleyrodidae )
Species-group placement and differential diagnosis. E. parvella -group (= E. pergandiella -group of authors, but distinct from E. longifasciata -group; also equivalent to Aleurodiphilus DeBach & Rose (1981)) . The group can be distinguished relatively easily by the following combination of characters: All tarsi 5-segmented; fore wing with a clear asetose (bare) area around the stigmal vein; marginal fringe of fore wing not longer than width of wing; each side lobe of the mesoscutum with 2 or more setae; parasitoids of Aleyrodidae . The other species currently in this group are the following:? E. acaudaleyrodis , E. americana , E. aseta , E. basicincta ,? E. citrofila , E. flavescens , E. gerlingi , E. guajavae , E . lanceolata, E. leucaenae ,? E. mineoi , E. nipponica , E. parvella , E. pergandiella , E. sueloderae (ending here emended from E. sueloderi ), E. tabacivora , E. telemachusi , E. tetraleurodis , E. trialeurodis , E. verticina . Of these, E. heratyi appears to be closest to E. trialeurodis Myartseva & Evans (2007) . It could also be confused with E. tetraleurodis Myartseva & Evans (2008) , and E. tabacivora Viggiani (1985) . Females of these species can be distinguished in the following table:
Species Fore wing infuscation F2 Ovipositor/mid tibia Colour of metasoma E. heratyi Slight , below stigmal vein only Without a sensillum 1.0 Mainly infuscate
E. tabacivora Extensive , below marginal vein Without a sensillum 0.9 Variable (dimorphic) E. tetraleurodis Extensive , below marginal vein With a sensillum 1.0 Completely yellow E. trialeurodis Slight , below stigmal vein only Without a sensillum 0.9 Completely yellow DNA sequence. The ribosomal 28S-D2-3 gene fragment was amplified and sequenced following non-destructive DNA extraction on the holotype female. Genomic DNA was extracted from each individual using a protocol largely based on that described in the DNeasy® Tissue Handbook provided by QIAGEN (www.qiagen.com). The following pair of primers was used for amplification:
Forward: D23F GAG AGT TCA AGA GTA CGT G
Reverse: 28Sb TCG GAA GGA ACC AGC TAC TA
Standard Polymerase Chain Reactions (PCR) were carried out in 25mL reactions containing 1x BIOTAQ Buffer, 1.5 mM MgCl2, 50 μM of each dNTP, 0.5 mM of each forward and reverse primer, 1.25 mM of BIOTAQ Taq polymerase and 5mL DNA extract. The PCR protocol was as follows: after the initial denaturation at 94ºC for 2 min the PCR reaction went through 40 cycles of 30 s at 94ºC, 30 s at 57ºC and 45 s at 72ºC; final extension was at 72ºC for 10 min.
Both strands of the PCR product were sequenced and the final sequence was produced by manually comparing both of them using SEQUENCHER version 3.0 (Gene Codes Corporation, Ann Arbor, MI, U.S.A.). This sequence has been deposited in GenBank under Accession No. HQ731041 View Materials .
The sequence was aligned with those of 28 other Encarsia sequences using Clustal W. The total of 29 Encarsia sequences, plus Coccophagoides moeris as an outgroup, were analysed using three different methods: parsimony (PAUP), Bayesian (MrBayes) and maximum likelihood (GARLI). In all three analyses, E. heratyi clustered together with three other parvella -group species.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |