Rhopilema esculentum Kishinouye, 1891
|
publication ID |
https://doi.org/ 10.5281/zenodo.5453269 |
|
persistent identifier |
https://treatment.plazi.org/id/03E287E8-6377-EC46-9526-FDFCFB88337B |
|
treatment provided by |
Valdenar |
|
scientific name |
Rhopilema esculentum Kishinouye, 1891 |
| status |
|
Rhopilema esculentum Kishinouye, 1891 View in CoL
( Fig. 1l View Fig )
Class: Scyphozoa
Family: Rhizostomatidae (Cuvier 1799)
Genus: Rhopilema (Haeckel 1880)
Description: This species has large scapulets, long manubrium and oral arms with numerous clubs or filaments usually with a large terminal club. The exumbrella is without ring canals ( Kramp 1961). The colouration of the umbrella and oral arms is reddish brown. This species may correspond to the “red type ” jellyfish as reported by Omori and Nakano (2001). The species has a perceptible sting. The exumbrella diameter of most specimens exceeds 450 mm.
Distribution and habitat: This edible species occurs seasonally in the Klang Strait and is harvested also in Sarawak ( Daud 1998). The distribution of R. esculentum includes Japan, China and the Bay of Korea. In China, the species is slightly reddish and also commercially harvested.
Molecular characterization
A total of 85 DNA sequences were obtained from the collected jellyfish samples, which comprised of 41 partial COI gene sequences (567- 588 bp); 24 partial 16S sequences (254-256 bp); and 20 ITS1 sequences (245-323 bp). The success of PCR and DNA sequencing varied among the species and the targeted region used; the COI, 16S and ITS1 sequences represented only 5 or 6 species. The final COI sequence alignment length was 588 bp which consisted of 76 taxa, inclusive of reference and outgroup species sequences. The COI sequence alignment that translates into 196 amino acid residues contained 301 variable sites and 283 parsimony informative sites. All the taxa showed similar nucleotide composition, with the average of T (thymine) = 35.0, C (cytosine) = 18.9, A (adenine) = 26.5 and G (guanine) = 19.5 percent. The final 16S sequence alignment length was 259 bp which consisted of 41 taxa, contained 156 variable sites and 142 parsimony informative sites. The nucleotide composition was similar across taxa, with an average of T = 27.4, C = 18.2, A = 35.8 and G = 18.6 percent. The final ITS1 sequence alignment length of 36 taxa was 394 bp, which contained 364 variable sites and 326 parsimony informative sites and many indels. The nucleotide composition was similar across taxa, with an average of T = 27.8, C = 22.7, A = 23.8 and G = 25.7 percent.
Phylogenetic analyses and molecular identification
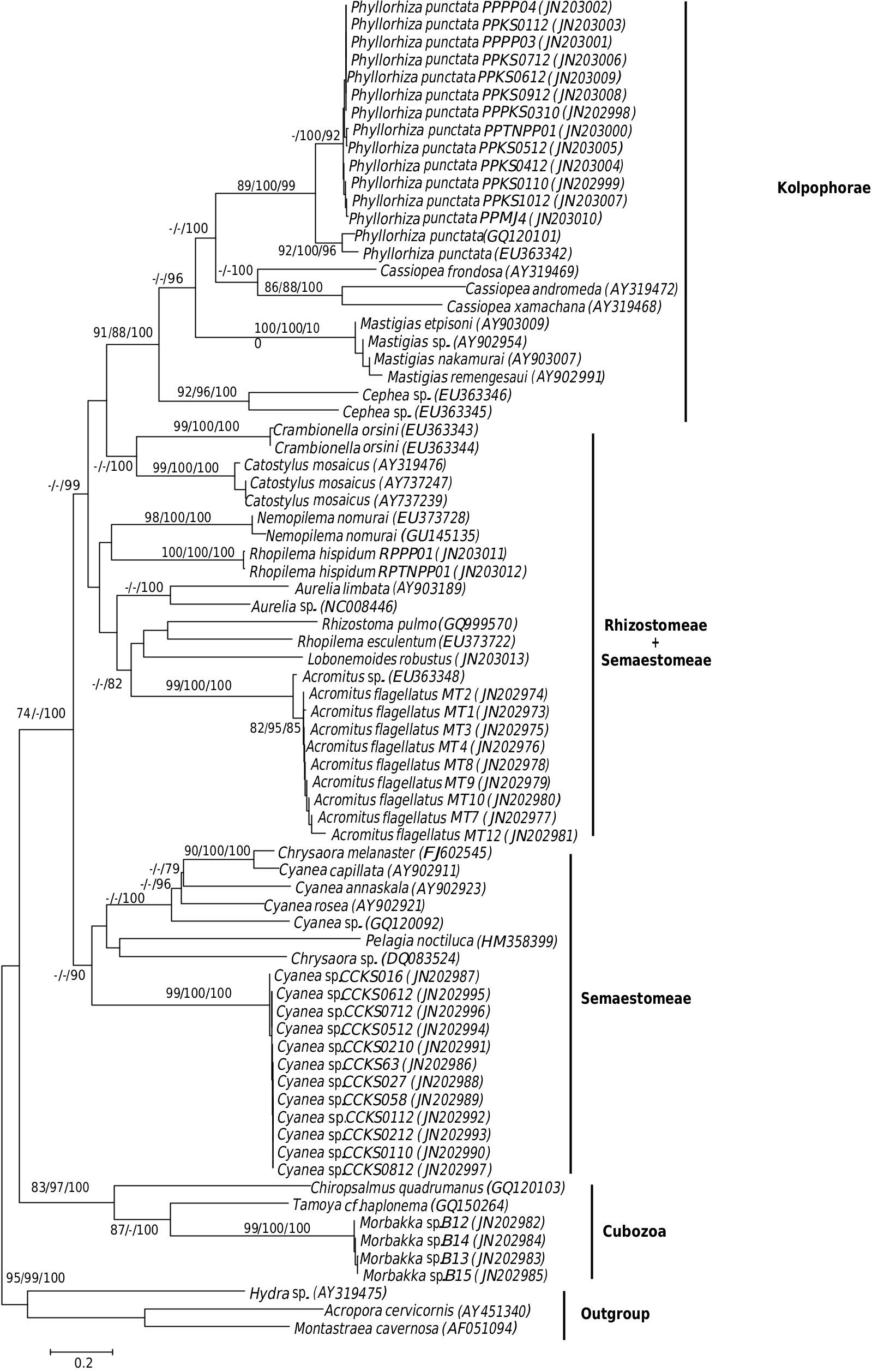
The NJ, MP and Bayesian analyses of the COI sequences produced congruent phylogenetic trees that are rooted with outgroups Anthozoa and Hydrozoa. The COI consensus phylogenetic tree of NJ, MP and Bayesian analyses shows separation of the six Malaysian jellyfish species ( Morbakka sp. , Rhopilema hispidum , Lobonemoides robustus , Acromitus flagellatus , Phyllorhiza punctata and the Malaysian Cyanea sp. ) with high bootstrap values and posterior probabilities at the species nodes ( Fig. 2 View Fig ).
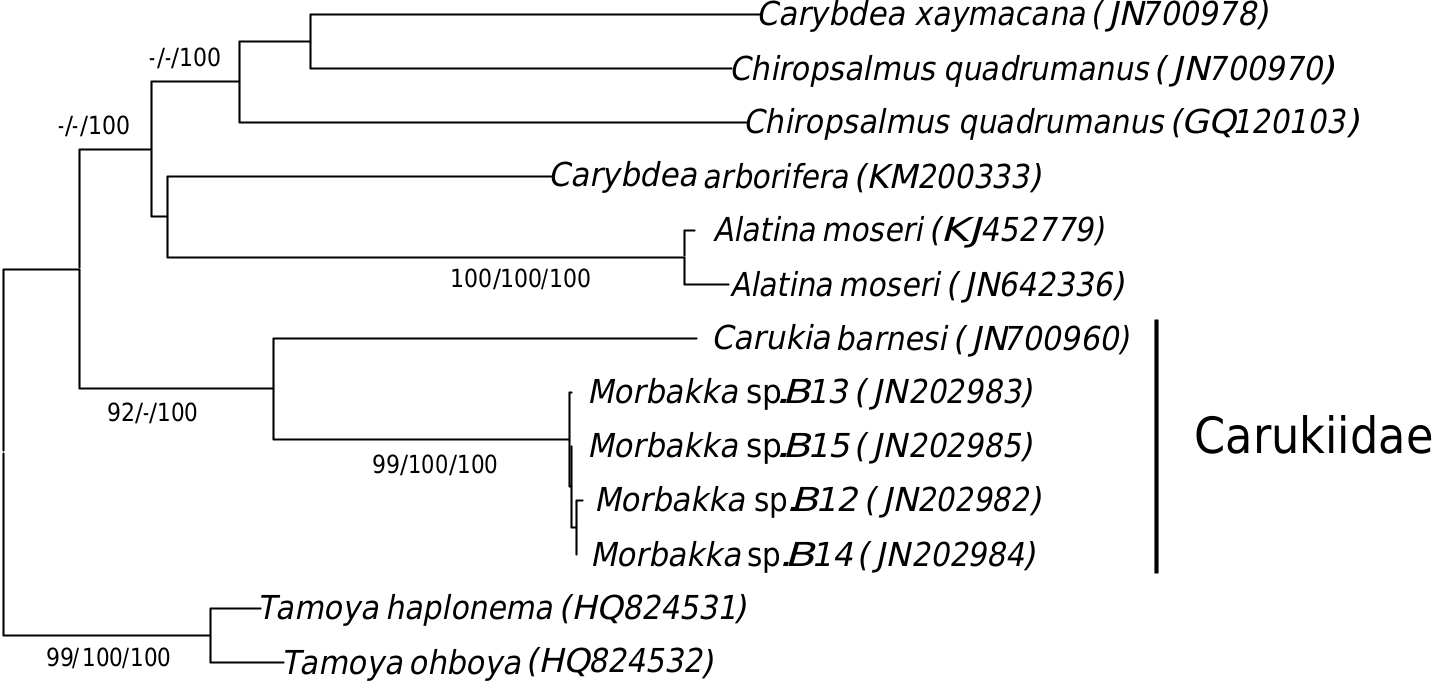
All of the mentioned species are also monophyletic and show affinity to four major clades. The Cubozoa clade contains sequences of the Malaysian Morbakka sp. that clustered with the reference sequences of Tamoya cf. haplonema [GenBank: GQ150264] a n d C h i r o p s a l m u s quadrumanus [GenBank: GQ120103] with average sequence diversity of 23.6% (± 0.3) and 28.8% (± 0.2), respectively. Although there were no reference COI sequences of Morbakka available for verification, the Malaysian Morbakka sp. sequences are strongly related to Carukia barnesi , suggesting it belongs to family Carukiidae ( Fig. 3 View Fig ). Coupled with its “rabbit ear”- like rhopaliar horns, it is highly possible that this box jellyfish belongs to the genus Morbakka .
Another clade identified is one that consists of the order Semaestomeae ( Aurelia sp. and Aurelia limbata ) and Rhizostomeae ( Rhopilema hispidum , Rhopilema esculentum , Acromitus flagellatus , Crambionella orsini , Catostylus mosaicus and Nemopilema nomurai ). The Malaysian R. hispidum sequences do not cluster with R. esculentum , but showed weak relationship with N. nomurai , another Rhizostomeae jellyfish. The identification of the Malaysian A. flagellatus is somewhat confirmed by its clustering with the Indian Acromitus sp. , with a sequence diversity of 2.0% (± 0.2). Besides having no available reference sequences for Lobonemoides robustus , the single sequence of this species showed no support for any specific sister group relationship with any other species within this Rhizostomeae and Semaestomeae mixed clade. The Malaysian Phyllorhiza punctata showed 8.9% (± 1.6) sequence diversity from the Australian Phyllorhiza punctata [GenBank: EU363342], inferring the possibility of being a cryptic species. P. punctata showed affinity to the Kolpophorae clade that also contained species members of Cassiopea , Mastigias and Cephea .
A distinctively Semaestomeae clade is also evident and comprises of Cyanea capillata , Cyanea rosea , Cyanea annaskala , Chrysaora sp. and Pelagia noctiluca . The Malaysian Cyanea sp. forms a different cluster from the reference sequences of C. capillata , C. annaskala and C. rosea , with an average sequence diversity of 25.0% (± 1.6). Although the Bayesian analysis appears to suggest paraphyly of Cyanea because the Chrysaora sequence clusters well with the Malaysian Cyanea sp. , this is not supported by other NJ and MP analysis.
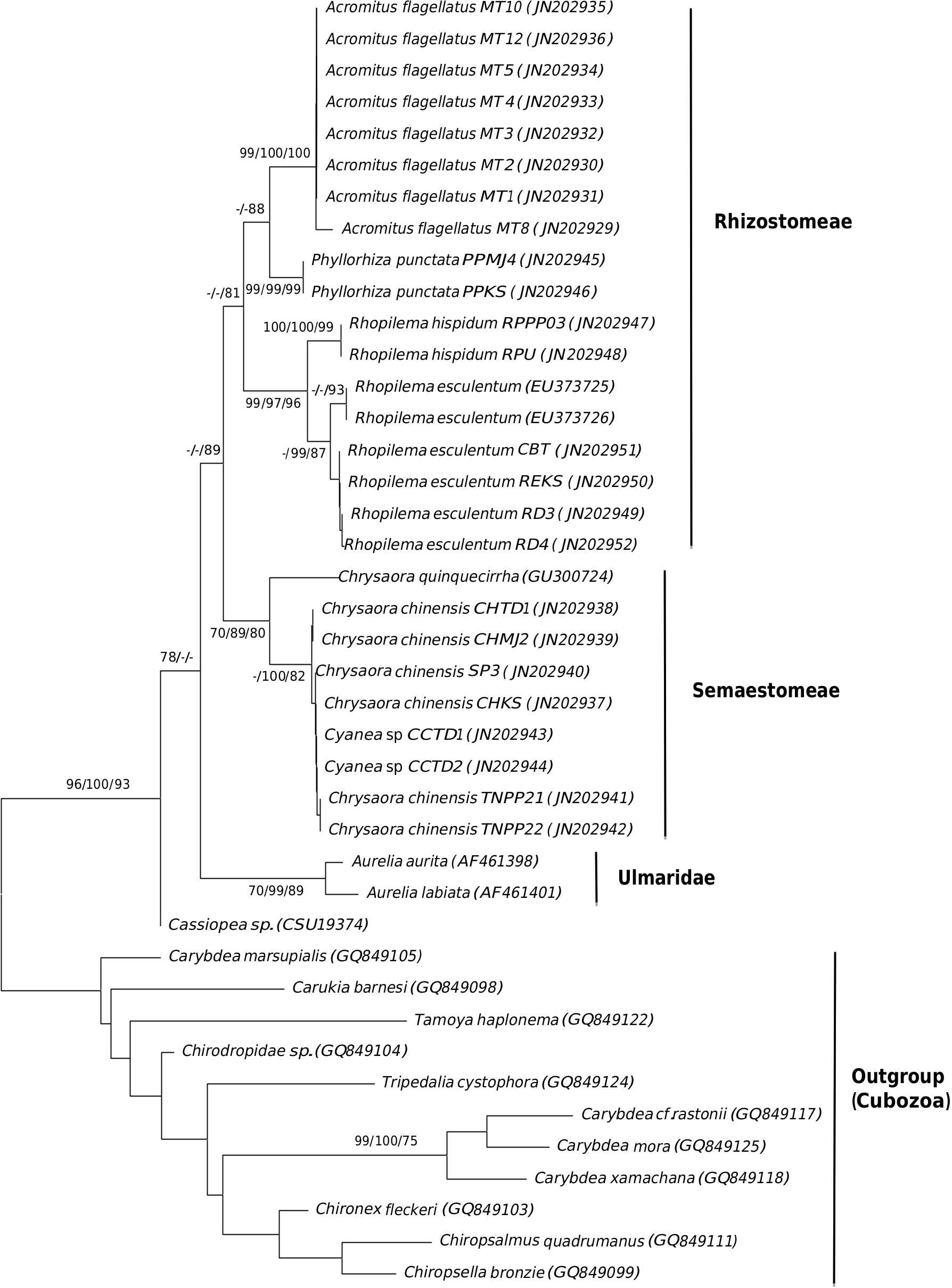
The 16S phylogeny which is rooted with reference sequences of Cubozoa, designated the Malaysian specimens of Chrysaora chinensis , the Malaysian Cyanea sp. , Rhopilema esculentum , Rhopilema hispidum , Acromitus flagellatus and Phyllorhiza punctata into two main clades; Rhizostomeae and Semaestomeae ( Fig. 4 View Fig ). The Rhizostomeae clade consists of Cassiopeia sp. , P. punctata , A. flagellatus , R. esculentum and R. hispidum . The tree shows close relatedness between R. esculentum and R. hispidum . The Malaysian R. esculentum clusters with R. esculentum from China [GenBank: EU373725 and EU373726] with an average sequence diversity of 4.6% (± 0.5). Within the Semaestomeae clade ( Chyrsaora quinquecirrha , C. chinensis and Cyanea sp. ), the Malaysian C. chinensis shows genetic affinity to the Malaysian Cyanea sp. rather than to other Chrysaora sp. (i.e. reference C. quinquecirrha ), with an average sequence diversity of 9.5% (± 0.5).
Congruencies between NJ, MP and Bayesian trees are also observed for the ITS1 region, which showed separation of the six Malaysian species ( Chiropsoides buitendijki , Chrysaora chinensis , Cyanea sp. , Lobonemoides robustus , Rhopilema esculentum and Rhopilema hispidum ) with high bootstrap values and posterior probabilities at the species node ( Fig. 5 View Fig ). These species are divided into five scyphozoan clades when rooted with the Cubozoa sequences (reference Chironex fleckeri and Malaysian C. buitendijki ). These clades represent the Rhizostomeae clade, three Cyanea clades and a mixed Semaestomeae and Rhizostomeae clade.
In the Rhizostomeae clade, the Malaysian Rhopilema esculentum clusters with the reference Rhopilema esculentum , thus confirming its identity. The Malaysian R. hispidum is a sister taxon to R. esculentum , consistent with being in the same genus. Although there were no reference sequences available to verify L. robustus , its distinct sequence showed weak affinity to the Rhizostomeae clade, which is consistent with its classification.
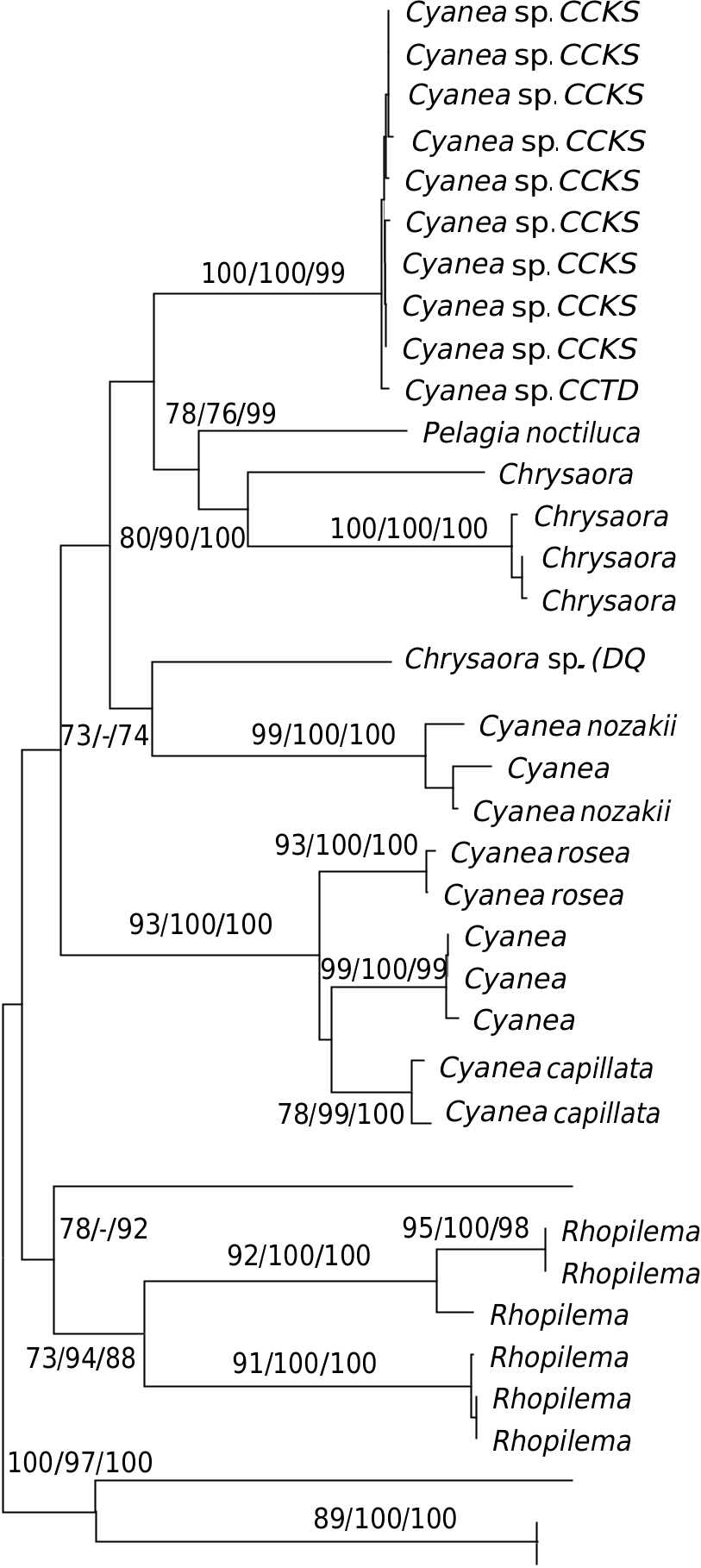
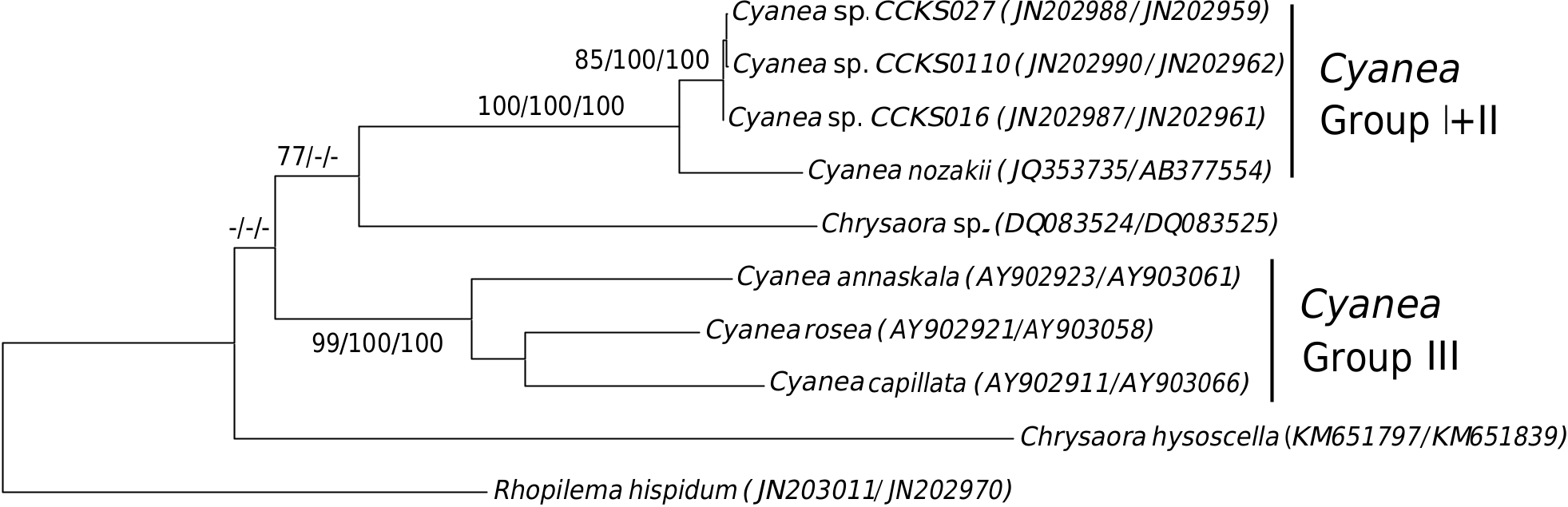
The ITS1 tree appears to show nonmonophyly of the genus Cyanea as there is distinct separation of the Malaysian Cyanea sp. , Cyanea nozakii and the Australian Cyanea species ( C. capillata , C. rosea and C. annaskala ) into different clades or groups ( Cyanea Group I-III). Although the genus Chrysaora appears paraphyletic, the evidence is weak and circumstantial. Sequences of Chrysaora chinensis [GenBank: JN202955- 57] from Malaysia are more closely related to Chrysaora fulgida [GenBank: HM348774], whereby both species are more related to Pelagia noctiluca [GenBank: HM348774] than to another reference sequence of Chrysaora sp. [GenBank: DQ083525]. The reference Chrysaora sp. is also related unexpectedly to the Japanese C. nozakii .
Phylogenetic analyses of concatenated COI and ITS1 sequences, failed to establish the monophyly of Cyanea and Chrysaora ( Fig. 6 View Fig ). The Australian Cyanea species remains separate from the Malaysian Cyanea sp. and Cyanea nozakii . Only Chrysaora sp. and Chrysaora hysoscella were available from GenBank for the total evidence molecular analysis. Unfortunately, their sister taxa relationship could not be determined due to poor resolution and support at the deep branches of the tree. Paraphyly of both Chrysaora and Cyanea remains inconclusive even though they appeared non -monophyletic.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.