Acorus calamus
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2022.113318 |
|
persistent identifier |
https://treatment.plazi.org/id/03D987ED-FF96-AE5E-FCCF-FF53B8DAFB16 |
|
treatment provided by |
Felipe |
|
scientific name |
Acorus calamus |
| status |
|
2.4. Analysis of sequences of terpene synthases from A. calamus View in CoL View at ENA
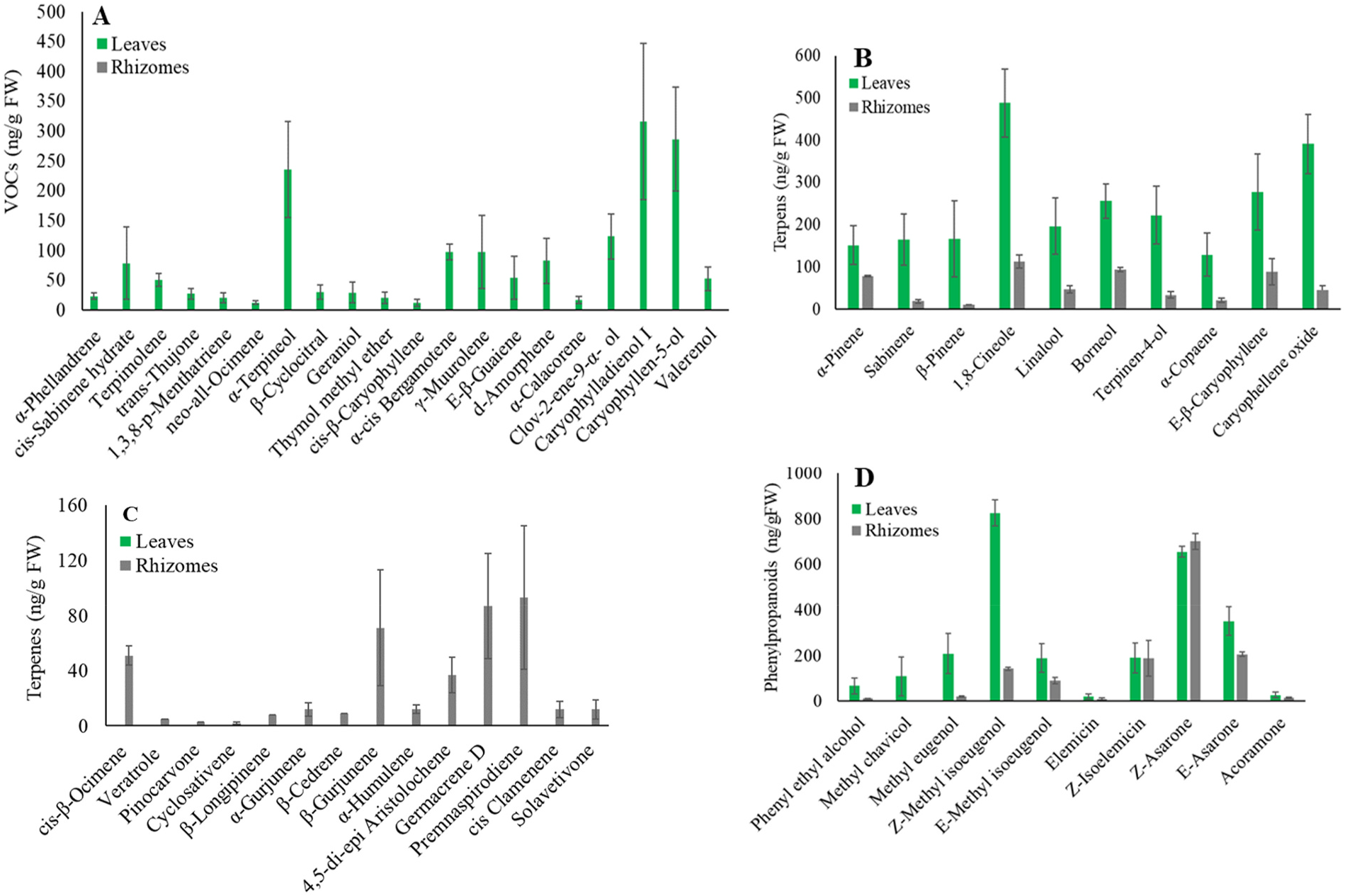
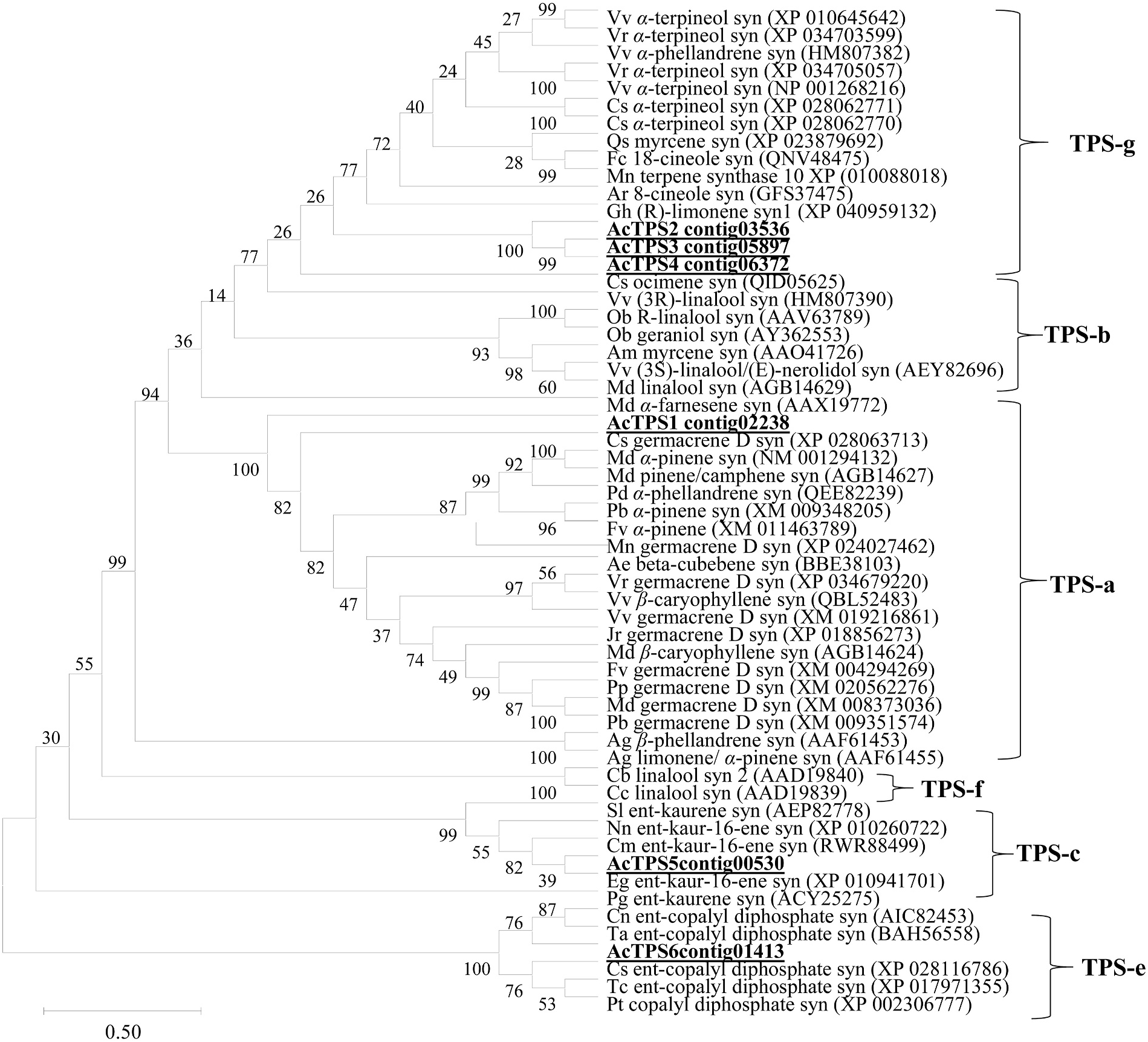
Based on our bioinformatics analysis, we were able to identify six contigs with sequence similarity to plant TPS genes, which were designated as Acorus calamus AcTPS 1 to AcTPS6 (Supplementary Fig. S1 View Fig ). From the sequence comparison with previously characterized TPSs from other plant species, the sequences of AcTPS1 to AcTPS6 appear to contain complete open reading frames (ORFs; Supplemental Fig. S1 View Fig ). A. calamus AcTPS 1 to AcTPS6 ORF sequences encoded 462, 494, 314, 339, 759, and 785 deduced amino acids, respectively. The calculated molecular weight of proteins encoded by these AcTPS genes ranged from 36.5 to 89.9 kDa (Supplementary Table S2). Among the 6 AcTPS genes, the TargetP-2.0 software (https://services.healthtech.dtu.dk/service.ph p?TargetP-2.0) predicted a plastidic localization for the proteins AcTPS2 and AcTPS6 with the putative amino-terminal extension of 32 and 39 amino acids, respectively, upstream of the RRx8W motif. A phylogenetic tree based on protein sequence comparisons with representative TPS sequences from other plant species indicated that the AcTPS1 protein belongs to the TPS-b clad, in which the majority of angiospermous monoterpene synthases reside ( Chen et al., 2011). AcTPS1 was found to have an identity with germacrene D synthase-like from Camellia sinensis , Vitis vinifera , and V. riparia (50% of amino acids identity) ( Fig. 4 View Fig ). The AcTPS2, AcTPS3, and AcTPS4 belong to the TPS-g clade, and the BLASTP analysis of these proteins with the NCBI protein database revealed that the α -terpineol synthase-like from V. riparia and Camellia sinensis have 50%, 52%, and 48% amino acid identity with AcTPS2, AcTPS3, and AcTPS4, respectively ( Fig. 4 View Fig ). Comparisons with representative TPS sequences from other plant species indicated that AcTPS-5 belongs to the clade TPS-c. The TPS sequence most similar to AcTPS5 is ent-kaur-16-ene synthase, from Elaeis guineensis (62% of amino acids) ( Fig. 4 View Fig ). As for AcTPS6 was found to belong to the clade TPS-e clade and had the highest sequence similarity of amino acids with ent -copalyl diphosphate synthase from C. sinensis (56% of amino acids identity) ( Fig. 4 View Fig ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |