Pempheris flavicycla
|
publication ID |
https://doi.org/10.11646/zootaxa.3887.3.5 |
|
publication LSID |
lsid:zoobank.org:pub:63968BBF-9C06-4A74-8093-0165770A6325 |
|
DOI |
https://doi.org/10.5281/zenodo.5694358 |
|
persistent identifier |
https://treatment.plazi.org/id/03D887FD-FF9C-FFCA-FF76-FF19FBEE3E52 |
|
treatment provided by |
Plazi |
|
scientific name |
Pempheris flavicycla |
| status |
|
Status of Pempheris flavicycla View in CoL and P. adusta
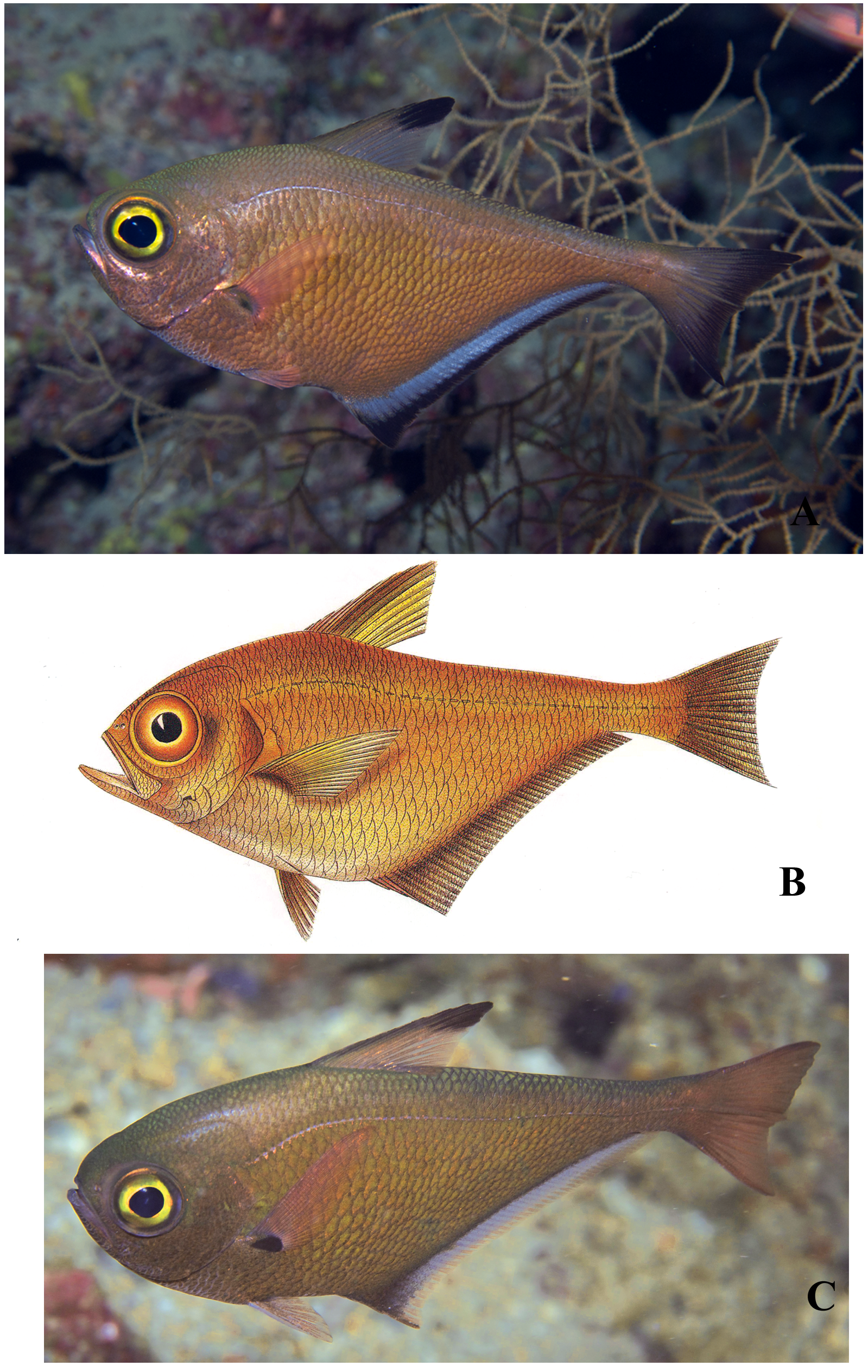
Pempheris adusta (type locality Ambon, on the Pacific side of Indonesia) is not a senior name for P. flavicycla of the Indian Ocean because of a pronounced difference in the markings on the anal fin. The salient dark marking of P. flavicycla is a prominent black border on the anal fin ( Fig. 1A View FIGURE 1. A ), which has led most past observers to mistake them for P. vanicolensis , a similar Pacific species that shares a prominent black border on the anal fin. Bleeker’s description of P. adusta makes no mention of a black border, nor is it shown on his painting ( Fig. 1 View FIGURE 1. A B). The holotype in the Leiden Museum also does not show any sign of a black border on the anal fin. In addition, P. flavicycla has an apical spot on the dorsal fin vs. an anterior dark band on Bleeker’s illustration.
Sweepers that match the Bleeker illustration are present in Indonesia: we identify the photograph taken by Mark Erdmann at the Raja Ampat Islands of West Papua, Indonesia as P. adusta ( Fig. 1 View FIGURE 1. A C). Plans to collect this species are underway. Bleeker specifically mentioned the fins as “ roseis ” with a black spot at the base of the pectoral fin, a dark brown anterior margin of the dorsal fin, a golden margin of the pupil, and his painting shows other diagnostic features such as the dark band at the base of the anal fin. Bleeker gave the anal-fin soft ray count as 42 (we get this count on Erdmann's photograph), and the lateral-line scale count as about 50. The holotype has 56 lateral-line scales on one side and 53 on the other (counted by Randall D. Mooi). We count about 58 on Erdmann's photograph. Note that Bleeker’s painting also shows two dark bars on the postorbital head: Martin van Oijen, curator of fishes at the museum in Leiden (now the Naturalis Biodiversity Center), reports that the marks are present only on the right side (evidently the artist thought they should also be shown on the left). We believe these marks are from original damage to the specimen.
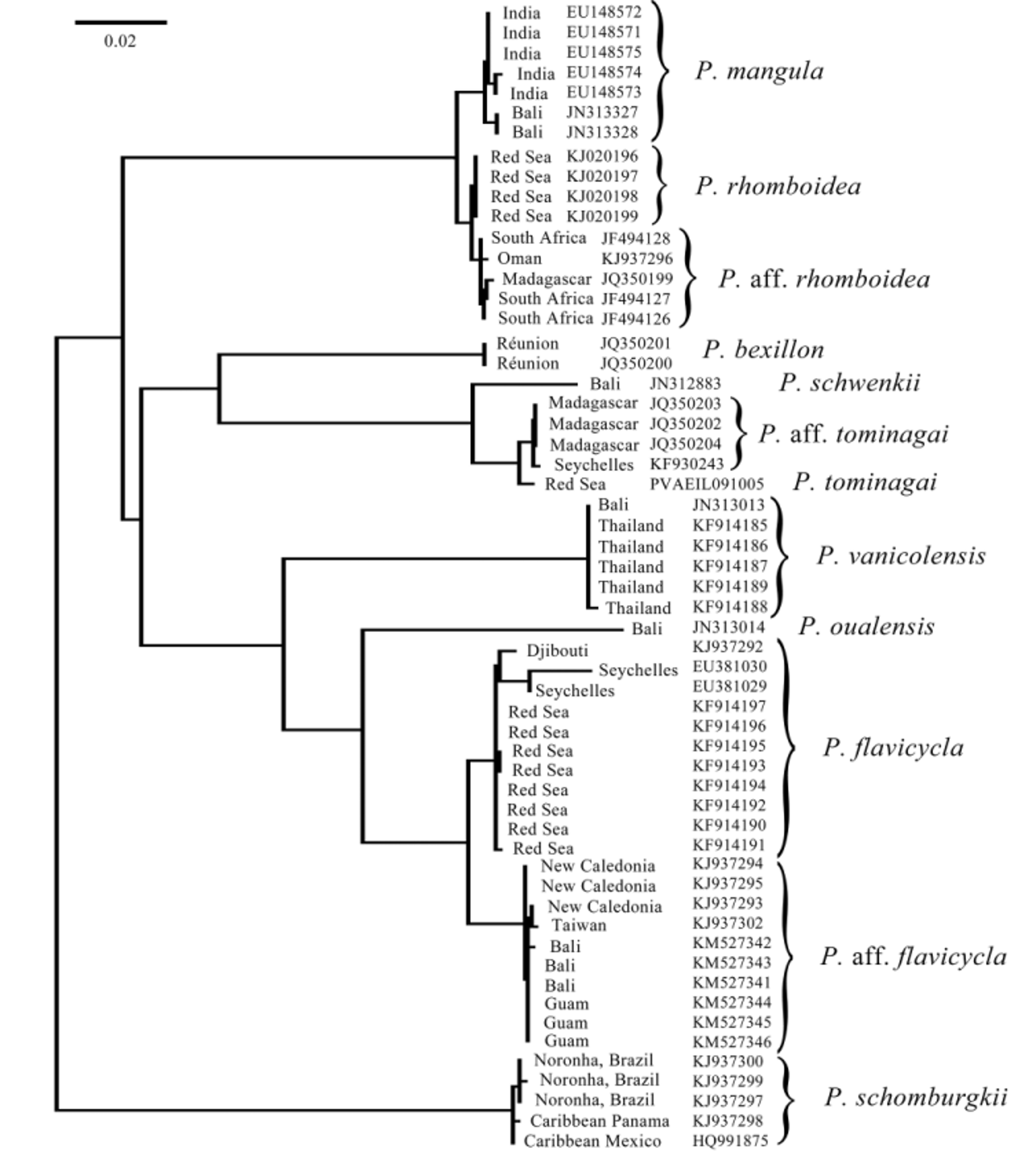
Despite this fundamental difference in markings, Koeda et al. (2014) concluded that the Red Sea species is P. adusta and then redescribed P. adusta to conform with this novel conclusion. They acknowledged in their diagnosis of “ P. adusta ” that the black border on the anal fin is absent in the Pacific: they say “usually none in Pacific” (p. 303), but add, notably without any evidence, that “some of the specimens from the Pacific have have [sic] the same coloration as fish from the Indian Ocean” (p. 305). It is especially significant that the only Pacific specimen they chose to illustrate, in their Fig. 1 View FIGURE 1. A d, has no black border on the anal fin. Then, inexplicably, they redescribe P. adusta “based on Indian Ocean specimens”, despite the obvious fact that Bleeker’s type locality of Ambon is from the Pacific Ocean. They switched oceans for the redescription undeterred by their own conclusion that there is a 0.4% divergence between the mtDNA 16S sequences for the Indian and Pacific Ocean lineages. Further confusing the argument, a typographical error allows the reader to conclude that the different ocean lineages were mixed: KAUM 31640 is listed in their phylogenetic tree (p. 308, Fig. 2 View FIGURE 2 ) as from “Iou-jima I., Thailand ”, but that sequence is from Japan (and URM-P 46441 is listed in the same tree as from “Tanega-shima I., Indonesia ”, but it is also from Japan; their Appendix 1, pp. 325 and 328). In fact, the two groups of lineages in the tree in their Fig. 2 View FIGURE 2 are mutually exclusive, with all Indian Ocean specimens sharing one haplotype that is displayed at a terminal node of the ML tree and all Pacific Ocean specimens sharing another haplotype that is ancestral to that of Indian Ocean specimens and therefore is found on the same branch as an inner node. This is exactly what can be expected if a short-enough sequence of a relatively slowly evolving gene is analysed for any two divergent evolutionary lineages.
The more broadly used mitochondrial gene COI, the marker for the Barcode of Life project with over 10,000 fish species sequenced at present (http://www.fishbol.org/), shows that Indian Ocean P. flavicycla and Pacific Ocean P. aff. flavicycla (“ P. adusta ” of the Pacific according to Koeda et al. 2014) are reciprocally monophyletic lineages 2.51% divergent (minimum interspecific distance by K2P, 2.46% pairwise; Fig. 2 View FIGURE 2 ), well within the norm for pairs of sibling species among coral reef fishes ( Ward et al. 2005). Our Pacific Ocean P. aff. flavicycla were collected from a very wide geographic range, i.e. Taiwan, Guam, Bali, and New Caledonia, yet form a tight monophyletic clade sister to the Indian Ocean P. flavicycla ( Fig. 2 View FIGURE 2 ). We confirmed that Koeda et al. ’s (2014) Pacific “ P. adusta ” is the same entity as our P. aff. flavicycla by sequencing 16S from our three Balinese specimens and the sequences are identical (GenBank numbers KM516206 View Materials – KM516208 View Materials ). Furthermore, our specimens of P. aff. flavicycla all share the essential features of their illustration of Pacific “ P. adusta ” (their Fig. 1 View FIGURE 1. A d), i.e. no black border on the anal fin and the yellow iris ringing the pupil of the eye.
Koeda et al. (2014) argued that the relatively low 0.4% divergence in the 16S sequence (and the paraphyly of one lineage) is actually evidence to synonymize the species, instead of considering that 16S may just have a slow substitution rate (especially compared to COI). Finding low divergences in mtDNA sequences should not trump morphological, meristic, or marking differences in assessing taxonomic questions. Indeed, numerous wellestablished reef-fish species complexes have very close mtDNA COI sequences, or even share identical haplotypes, due to incomplete lineage sorting (not enough time after speciation for mitochondrial haplotypes to become completely mutually exclusive) or due to rare hybridization (Victor, in press).
As a comparison, Koeda et al. (2014) cited that P. oualensis has a much higher sequence variation of 2.8% within the species (it should be noted that comparative sequence variation is not particularly useful for taxonomic questions). Nevertheless, that high variation is based on misidentifications in databases: nominal P. oualensis of the Pacific Ocean comprise several species, some long-established, such as P. otaitensis Cuvier 1831 , and some new species which are being presently described by us. Koeda et al. (2014) unwittingly included sequences of apparent P. otaitensis from French Polynesia in their phylogenetic tree (p. 308, Fig. 2 View FIGURE 2 ) as P. oualensis (USNM 402187, 402188 and four uncatalogued USNM specimens from French Polynesia; note that none of the six sequences are present on GenBank and the USNM catalog lists the collection location for USNM 402187, 402188 as Mohotane Island, Marquesas Islands, not Mururoa Atoll (Tuamotu Archipelago) as listed in their Appendix 2). Unfortunately, they added to the existing confusion regarding species designations and related sequence information in public databases by depositing in GenBank a sequence ( AB773862 View Materials ) that is identical to their deposited sequences for “ Pempheris adusta ” ( AB702676 View Materials and AB702677 View Materials ) for nine specimens of “ Pempheris mangula ” (i.e. P. rhomboidea ) from the Mediterranean (HUJ 19999, HUJ 20099 (No.1–4) and SAIAB 81068 (No.1–4)). This is certainly not the sequence they used in their analysis of the partial 16S ribosomal DNA.
These errors and misidentifications by Koeda et al. (2014), as well as the missed public DNA sequences for the type locality of P. mangula (discussed below), highlight the risk of ignoring large databases such as the Barcode Database (BOLD), a frequent oversight by some using molecular taxonomy, who tend to focus on the gene marker they favor rather than the markers with the most complete or optimal data, or, ideally, sets of multiple markers, both nuclear and mitochondrial (Victor, in press). Koeda et al. (2014) used mostly sequences obtained from material in museums from Japan and were apparently unaware of the public sequences in the COI database that would perhaps have changed their conclusions.
Koeda et al. (2014) did not note an error in the paper by Randall et al. (2013) describing Pempheris flavicycla ; they might have questioned the lateral-line scale count of 57 given to a syntype of P. vanicolensis from the Pacific, when Table 6 of Randall et al. (2013) shows that count fits very well in the range of P. flavicycla , not P. vanicolensis . By explanation, Randall was sent on loan one of four syntypes of P. vanicolensis from the Múseum national d’Histoire naturelle in Paris in 1989. After noting that most of the scales are missing, he gave an approximate lateral-line scale count of 57, in actuality an undercount. Mooi & Randall (2014: Table 1) correctly listed the lateral-line scale counts of the four syntypes of P. vanicolensis as 63 and 64.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |