Furfurilactobacillus, Zheng & Wittouck & Salvetti & Franz & Harris & Mattarelli & O’Toole & Pot & Vandamme & Walter & Watanabe & Wuyts & Felis & Gänzle & Lebeer, 2020
|
publication ID |
https://doi.org/10.1099/ijsem.0.004107 |
|
DOI |
https://doi.org/10.5281/zenodo.4728701 |
|
persistent identifier |
https://treatment.plazi.org/id/03A8D903-D221-026F-FC95-FEA852E53726 |
|
treatment provided by |
Valdenar (2021-04-29 20:05:09, last updated by Guilherme 2025-02-06 14:43:30) |
|
scientific name |
Furfurilactobacillus |
| status |
gen. nov. |
DESCRIPTIONOF FURFURILACTOBACILLUS GEN. NOV.
Furfurilactobacillus (Fur.fu.ri.lac.to.ba.cil’lus. L. masc. n. furfur bran, relating to the origin of furfurilactobacilli from cereal fermentations; N.L. masc. n. Lactobacillus a bacterial genus name; N.L. masc. n. Furfurilactobacillus a lactobacillus from bran).
Heterofermentative and aerotolerant. Growth is observed at 15 and 37°C but not at 45 °C. The two species in the genus with genome sequences available have a genome size of 2.9–3.0 Mbp and a mol% G+C content of DNA of 43–44 %. Species in thegenus were isolated from sourdough or spoiled beer and have an exceptional capacity to metabolize phenolic compounds [ 240, 241]. The ecology of the genus remains largely unexplored but appears to be similar to the nomadic lifestyle of L. plantarum .
Aphylogenetic tree on the basis of 16S rRNA genes of all species in the genus Lactiplantibacillus is provided in Figure S6L View Fig .
The type species of the genus is Furfurilactobacillus rossiae comb. nov.; Furfurilactobacillus was previously referred to as L. rossiae group.
241. Gaur G, J-H O, Filannino P, Gobbetti M, van Pijkeren J-P et al. Genetic determinants of hydroxycinnamic acid metabolism in heterofermentative lactobacilli. Appl Environ Microbiol 2020; 86: e 02461 - 19.
240. Ripari V, Bai Y, Ganzle MG. Metabolism of phenolic acids in whole wheat and rye malt sourdoughs. Food Microbiol 2019; 77: 43 - 51.
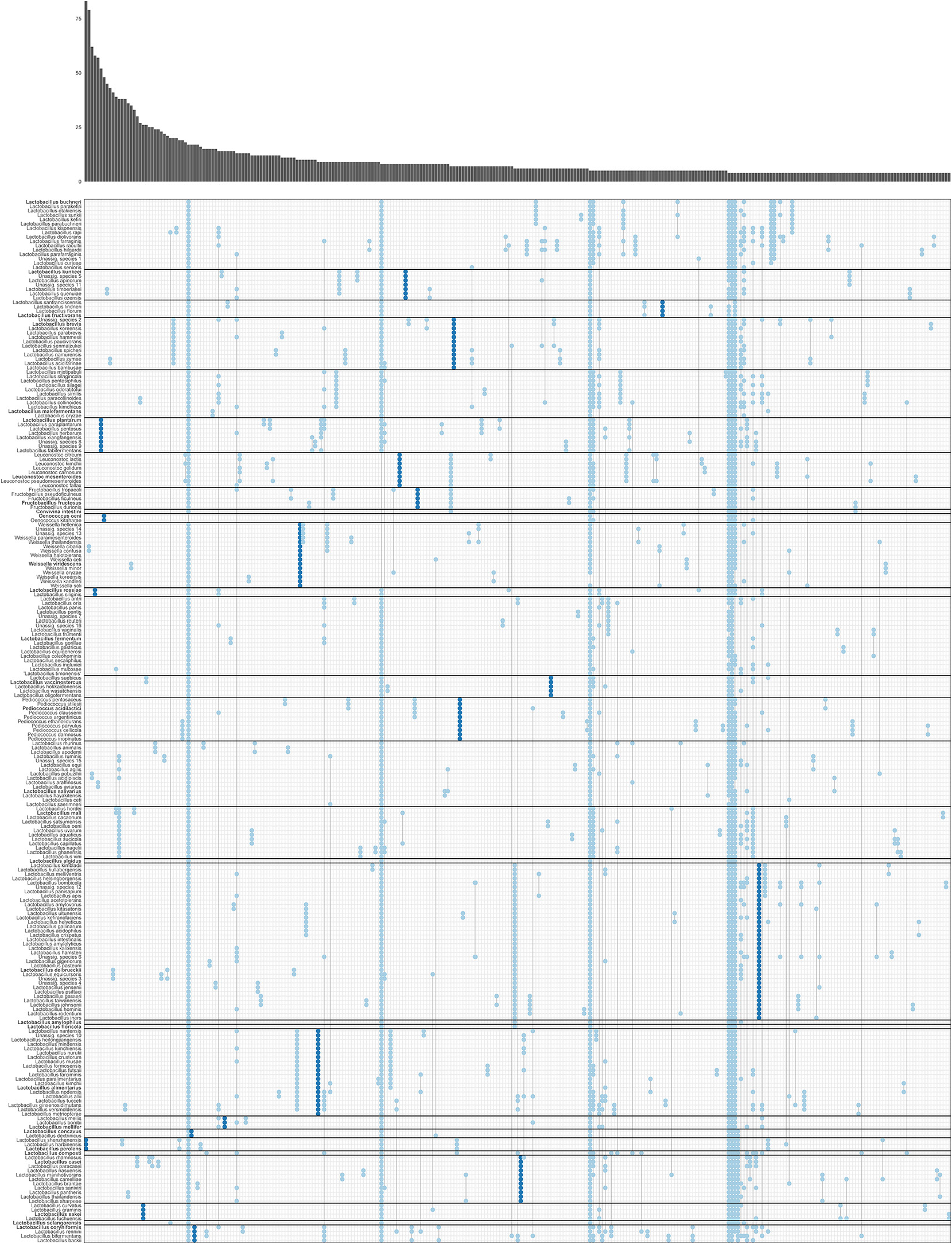
Fig. 6. Gene family presence/absence patterns in Lactobacillaceae and Leuconostocaceae. Each column represents a gene family presence/absence pattern in species of Lactobacillaceae and Leuconostoaceae, where presence is indicated with a dot. The absolute number of gene families that conform to each pattern is visualized in the marginal bar plot at the top. Separations between phylogroups are indicated with horizontal black lines. We defined genes that were present in all genomes of a clade and in none of the genomes outside of that clade as ‘signature genes’ (dark blue); other genes are shown in light blue. Only presence/absence patterns followed by four or more gene families are shown. Patterns of presence in a single species or all species are not shown. Unassigned species are clusters of closely related genomes which could not be assigned to a known species due to low whole-genome similarity to a type strain and/or low 16S rRNA similarity to a type strain.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
1 (by valdenar, 2021-04-29 20:05:09)
2 (by ExternalLinkService, 2021-04-29 21:47:49)
3 (by valdenar, 2021-04-30 14:27:18)
4 (by valdenar, 2021-04-30 14:46:18)
5 (by ExternalLinkService, 2021-04-30 19:06:52)
6 (by valdenar, 2021-04-30 19:23:20)
7 (by ExternalLinkService, 2022-01-29 07:26:42)
8 (by valdenar, 2022-06-14 18:02:19)
9 (by plazi, 2023-11-02 08:01:04)