Oroxylum indicum subsp. root, (L.) Kurz
|
publication ID |
https://doi.org/10.1016/j.phytochem.2018.09.013 |
|
DOI |
https://doi.org/10.5281/zenodo.10570093 |
|
persistent identifier |
https://treatment.plazi.org/id/03881B76-8359-7F01-FCCD-ECEE8315F98B |
|
treatment provided by |
Felipe |
|
scientific name |
Oroxylum indicum subsp. root |
| status |
|
2.8. Identification and phylogenetic analysis of cytochrome P 450s (CYP450s) putatively involved in biosynthesis of specialized flavonoids like baicalein in O. indicum root
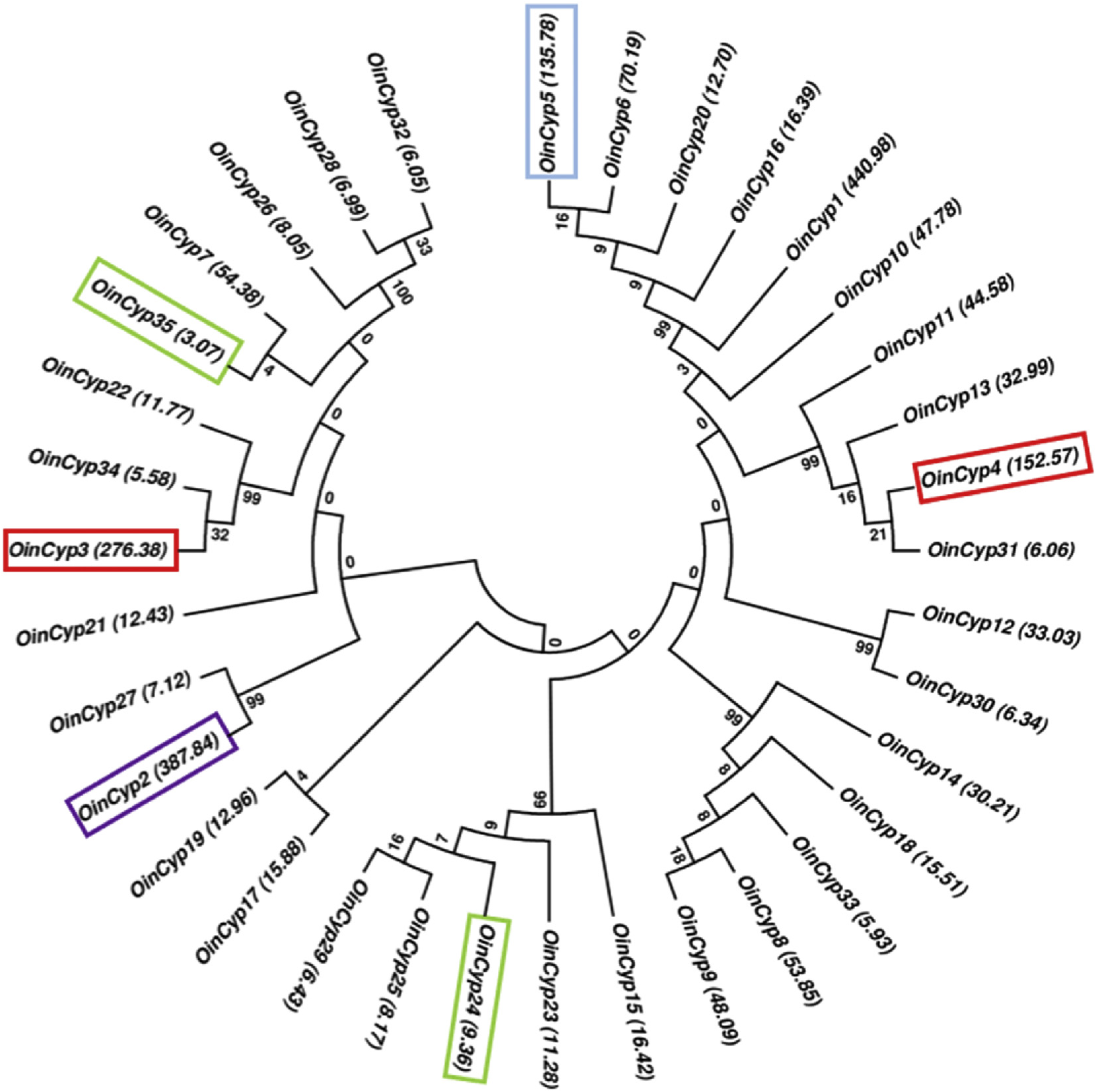
Command line BLAST was performed on the O. indicum transcriptome using flavone synthase II, flavone-6-hydroxylase, glucuronidase and transferase genes reported from Scutellaria baicalensis and shown to be involved in the biosynthesis of specialized flavonoids. This led to the identification of the O. indicum homologs of those genes as listed (Table 7). Additionally, 164 transcripts annotated as CYP 4 50s from O. indicum transcriptome were analyzed (Supplementary Table 4 View Table 4 .) Of these, the ones with high FPKM value (more than 5.5), were screened for identification of full length CYP450 ORFs from the O. indicum root transcriptome. A total of 31 genes ( OinCyp450) were identified ( Table 8 View Table 8 ). The phylogenetic tree of these OinCyp 4 50s indicated a high diversity of CYP450s in the O. indicum root ( Fig. 5 View Fig ). At least five OinCyps showed a high degree of similarity to biochemically well characterized enzymes from S. baicalensis namely, flavones synthase II, baicalin-ss-D-glucuronidase, flavone-6-hydroxylase, baicalin-7-O-glucuronosyltransferase ( Liu et al., 2015; Zhao et al., 2016, 2018). These enzymes catalyze the biosynthesis of specialized flavonoids like baicalin and wogonin through a root specific alternative pathway (Zhao et al., 2016).
The OinCyp3 and OinCyp4 were abundant in roots of O. indicum and showed a high degree of similarity with flavone synthase II (CYP93B; Zhao et al., 2016) along with five more OinCyps ( Fig. 5 View Fig ). Moreover, OinCyp2 (with similarity to S. baicalensis baicalin-ss-D-glucuronidase) clustered close to OinCyp27. OinCyp2 and OinCyp35 (both flavone-6- hydroxylase) clustered away from each other forming two different OinCyp clades ( Fig. 5 View Fig ). OinCyp5 (with similarity to S. baicalensis baicalin-7-O-glucuronosyl transferase) was highly abundant in O. indicum roots and formed a cluster with at least five more OinCyps. Based on the phylogenetic analysis of the OinCyps, it can be hypothesized that they are involved in the specialized flavonoid biosynthesis in O. indicum roots. Further, the functional validation of OinCyps may highlight their specific biochemical roles in metabolite biosynthesis in O. indicum .
Phylogenetic variations in Cyp450 genes play a central role in evolution of metabolic complexity ( Bak et al., 2011). Cyp9 8 A and its duplicated sister genes evolved functional diversification in phenolic and flavonoid biosynthesis pathways ( Liu et al., 2016). The flavonoid biosynthesis pathway is known to involve the formation of metabolons or protein complexes ( Bassard et al., 2017). It has been demonstrated that Cyp450s like flavone synthase and cinnamate 4-hydroxylase are ER membrane anchored enzymes and provide a nucleation platform for the assembly of other soluble enzymes like chalcone synthase, chalcone isomerase and reductase. Together, these complexes drive the flavonoid metabolism ( Dastmalchi et al., 2016) and plant specific diversity in a flavonoid metabolon organization ( Fujino et al., 2018). Detailed functional validation of OinCyp diversity will open up their precise biochemical roles and novelty in specialized flavonoid metabolism in O. indicum .
| P |
Museum National d' Histoire Naturelle, Paris (MNHN) - Vascular Plants |
| A |
Harvard University - Arnold Arboretum |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |